-
- Page not found -
-The page you requested cannot be found (perhaps it was moved or renamed).
-You may want to try searching to find the page's new location, or use -the table of contents to find the page you are looking for.
-
-
-
-
The page you requested cannot be found (perhaps it was moved or renamed).
-You may want to try searching to find the page's new location, or use -the table of contents to find the page you are looking for.
- -
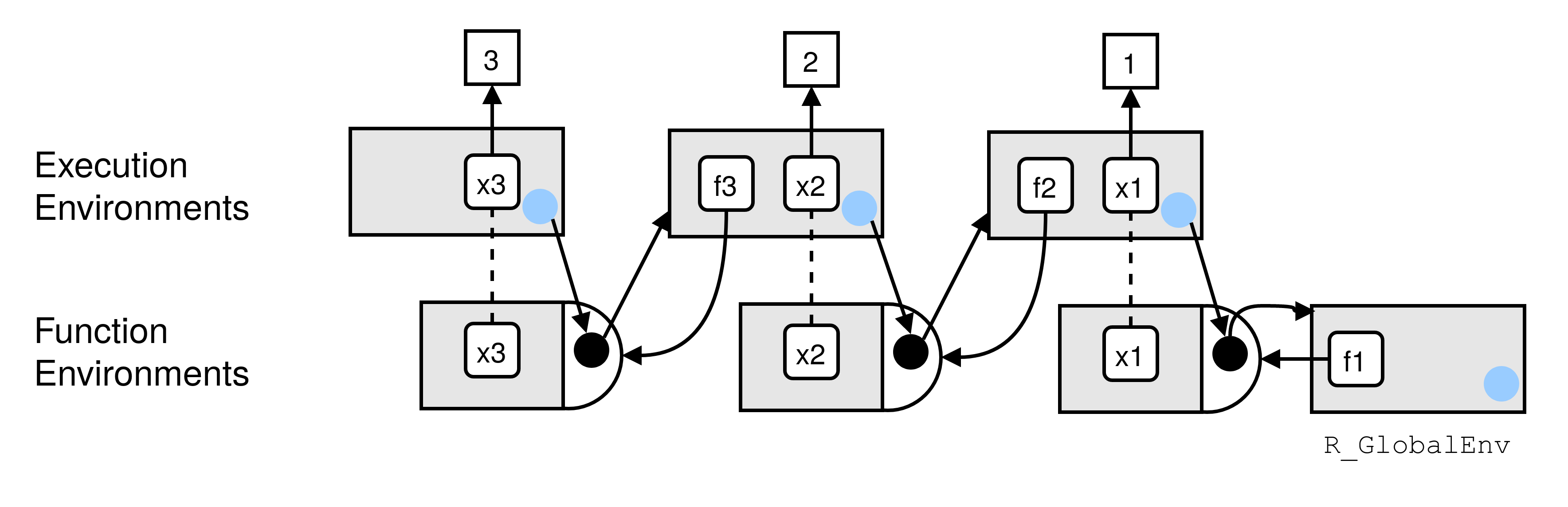
-**A2.** Creating the environment illustrated in the picture:
-
-
-```r
-library(rlang)
-
-e <- env()
-e$loop <- e
-env_print(e)
-#>
-
-**A2.** Creating the environment illustrated in the picture:
-
-
-```r
-library(rlang)
-
-e <- env()
-e$loop <- e
-env_print(e)
-#>  -
-**A3.** Creating the specified environment:
-
-
-```r
-e1 <- env()
-e2 <- env()
-
-e1$loop <- e2
-e2$deloop <- e1
-
-# following should be the same
-lobstr::obj_addrs(list(e1, e2$deloop))
-#> [1] "0x1083b4018" "0x1083b4018"
-lobstr::obj_addrs(list(e2, e1$loop))
-#> [1] "0x108409438" "0x108409438"
-```
-
-**Q4.** Explain why `e[[1]]` and `e[c("a", "b")]` don't make sense when `e` is an environment.
-
-**A4.** An environment is a non-linear data structure, and has no concept of ordered elements. Therefore, indexing it (e.g. `e[[1]]`) doesn't make sense.
-
-Subsetting a list or a vector returns a subset of the underlying data structure. For example, subsetting a vector returns another vector. But it's unclear what subsetting an environment (e.g. `e[c("a", "b")]`) should return because there is no data structure to contain its returns. It can't be another environment since environments have reference semantics.
-
-**Q5.** Create a version of `env_poke()` that will only bind new names, never re-bind old names. Some programming languages only do this, and are known as [single assignment languages](https://en.wikipedia.org/wiki/Assignment_(computer_science)#Single_assignment).
-
-**A5.** Create a version of `env_poke()` that doesn't allow re-binding old names:
-
-
-```r
-env_poke2 <- function(env, nm, value) {
- if (env_has(env, nm)) {
- abort("Can't re-bind existing names.")
- }
-
- env_poke(env, nm, value)
-}
-```
-
-Making sure that it behaves as expected:
-
-
-```r
-e <- env(a = 1, b = 2, c = 3)
-
-# re-binding old names not allowed
-env_poke2(e, "b", 4)
-#> Error in `env_poke2()`:
-#> ! Can't re-bind existing names.
-
-# binding new names allowed
-env_poke2(e, "d", 8)
-e$d
-#> [1] 8
-```
-
-Contrast this behavior with the following:
-
-
-```r
-e <- env(a = 1, b = 2, c = 3)
-
-e$b
-#> [1] 2
-
-# re-binding old names allowed
-env_poke(e, "b", 4)
-e$b
-#> [1] 4
-```
-
-**Q6.** What does this function do? How does it differ from `<<-` and why might you prefer it?
-
-
-```r
-rebind <- function(name, value, env = caller_env()) {
- if (identical(env, empty_env())) {
- stop("Can't find `", name, "`", call. = FALSE)
- } else if (env_has(env, name)) {
- env_poke(env, name, value)
- } else {
- rebind(name, value, env_parent(env))
- }
-}
-rebind("a", 10)
-#> Error: Can't find `a`
-a <- 5
-rebind("a", 10)
-a
-#> [1] 10
-```
-
-**A6.** The downside of `<<-` is that it will create a new binding if it doesn't exist in the given environment, which is something that we may not wish:
-
-
-```r
-# `x` doesn't exist
-exists("x")
-#> [1] FALSE
-
-# so `<<-` will create one for us
-{
- x <<- 5
-}
-
-# in the global environment
-env_has(global_env(), "x")
-#> x
-#> TRUE
-x
-#> [1] 5
-```
-
-But `rebind()` function will let us know if the binding doesn't exist, which is much safer:
-
-
-```r
-rebind <- function(name, value, env = caller_env()) {
- if (identical(env, empty_env())) {
- stop("Can't find `", name, "`", call. = FALSE)
- } else if (env_has(env, name)) {
- env_poke(env, name, value)
- } else {
- rebind(name, value, env_parent(env))
- }
-}
-
-# doesn't exist
-exists("abc")
-#> [1] FALSE
-
-# so function will produce an error instead of creating it for us
-rebind("abc", 10)
-#> Error: Can't find `abc`
-
-# but it will work as expected when the variable already exists
-abc <- 5
-rebind("abc", 10)
-abc
-#> [1] 10
-```
-
-## Recursing over environments (Exercises 7.3.1)
-
-**Q1.** Modify `where()` to return _all_ environments that contain a binding for `name`. Carefully think through what type of object the function will need to return.
-
-**A1.** Here is a modified version of `where()` that returns _all_ environments that contain a binding for `name`.
-
-Since we anticipate more than one environment, we dynamically update a list each time an environment with the specified binding is found. It is important to initialize to an empty list since that signifies that given binding is not found in any of the environments.
-
-
-```r
-where <- function(name, env = caller_env()) {
- env_list <- list()
-
- while (!identical(env, empty_env())) {
- if (env_has(env, name)) {
- env_list <- append(env_list, env)
- }
-
- env <- env_parent(env)
- }
-
- return(env_list)
-}
-```
-
-Let's try it out:
-
-
-```r
-where("yyy")
-#> list()
-
-x <- 5
-where("x")
-#> [[1]]
-#>
-
-**A3.** Creating the specified environment:
-
-
-```r
-e1 <- env()
-e2 <- env()
-
-e1$loop <- e2
-e2$deloop <- e1
-
-# following should be the same
-lobstr::obj_addrs(list(e1, e2$deloop))
-#> [1] "0x1083b4018" "0x1083b4018"
-lobstr::obj_addrs(list(e2, e1$loop))
-#> [1] "0x108409438" "0x108409438"
-```
-
-**Q4.** Explain why `e[[1]]` and `e[c("a", "b")]` don't make sense when `e` is an environment.
-
-**A4.** An environment is a non-linear data structure, and has no concept of ordered elements. Therefore, indexing it (e.g. `e[[1]]`) doesn't make sense.
-
-Subsetting a list or a vector returns a subset of the underlying data structure. For example, subsetting a vector returns another vector. But it's unclear what subsetting an environment (e.g. `e[c("a", "b")]`) should return because there is no data structure to contain its returns. It can't be another environment since environments have reference semantics.
-
-**Q5.** Create a version of `env_poke()` that will only bind new names, never re-bind old names. Some programming languages only do this, and are known as [single assignment languages](https://en.wikipedia.org/wiki/Assignment_(computer_science)#Single_assignment).
-
-**A5.** Create a version of `env_poke()` that doesn't allow re-binding old names:
-
-
-```r
-env_poke2 <- function(env, nm, value) {
- if (env_has(env, nm)) {
- abort("Can't re-bind existing names.")
- }
-
- env_poke(env, nm, value)
-}
-```
-
-Making sure that it behaves as expected:
-
-
-```r
-e <- env(a = 1, b = 2, c = 3)
-
-# re-binding old names not allowed
-env_poke2(e, "b", 4)
-#> Error in `env_poke2()`:
-#> ! Can't re-bind existing names.
-
-# binding new names allowed
-env_poke2(e, "d", 8)
-e$d
-#> [1] 8
-```
-
-Contrast this behavior with the following:
-
-
-```r
-e <- env(a = 1, b = 2, c = 3)
-
-e$b
-#> [1] 2
-
-# re-binding old names allowed
-env_poke(e, "b", 4)
-e$b
-#> [1] 4
-```
-
-**Q6.** What does this function do? How does it differ from `<<-` and why might you prefer it?
-
-
-```r
-rebind <- function(name, value, env = caller_env()) {
- if (identical(env, empty_env())) {
- stop("Can't find `", name, "`", call. = FALSE)
- } else if (env_has(env, name)) {
- env_poke(env, name, value)
- } else {
- rebind(name, value, env_parent(env))
- }
-}
-rebind("a", 10)
-#> Error: Can't find `a`
-a <- 5
-rebind("a", 10)
-a
-#> [1] 10
-```
-
-**A6.** The downside of `<<-` is that it will create a new binding if it doesn't exist in the given environment, which is something that we may not wish:
-
-
-```r
-# `x` doesn't exist
-exists("x")
-#> [1] FALSE
-
-# so `<<-` will create one for us
-{
- x <<- 5
-}
-
-# in the global environment
-env_has(global_env(), "x")
-#> x
-#> TRUE
-x
-#> [1] 5
-```
-
-But `rebind()` function will let us know if the binding doesn't exist, which is much safer:
-
-
-```r
-rebind <- function(name, value, env = caller_env()) {
- if (identical(env, empty_env())) {
- stop("Can't find `", name, "`", call. = FALSE)
- } else if (env_has(env, name)) {
- env_poke(env, name, value)
- } else {
- rebind(name, value, env_parent(env))
- }
-}
-
-# doesn't exist
-exists("abc")
-#> [1] FALSE
-
-# so function will produce an error instead of creating it for us
-rebind("abc", 10)
-#> Error: Can't find `abc`
-
-# but it will work as expected when the variable already exists
-abc <- 5
-rebind("abc", 10)
-abc
-#> [1] 10
-```
-
-## Recursing over environments (Exercises 7.3.1)
-
-**Q1.** Modify `where()` to return _all_ environments that contain a binding for `name`. Carefully think through what type of object the function will need to return.
-
-**A1.** Here is a modified version of `where()` that returns _all_ environments that contain a binding for `name`.
-
-Since we anticipate more than one environment, we dynamically update a list each time an environment with the specified binding is found. It is important to initialize to an empty list since that signifies that given binding is not found in any of the environments.
-
-
-```r
-where <- function(name, env = caller_env()) {
- env_list <- list()
-
- while (!identical(env, empty_env())) {
- if (env_has(env, name)) {
- env_list <- append(env_list, env)
- }
-
- env <- env_parent(env)
- }
-
- return(env_list)
-}
-```
-
-Let's try it out:
-
-
-```r
-where("yyy")
-#> list()
-
-x <- 5
-where("x")
-#> [[1]]
-#>  -
-**Q3.** Write an enhanced version of `str()` that provides more information about functions. Show where the function was found and what environment it was defined in.
-
-**A3.** To write the required function, we can first re-purpose the `fget()` function we wrote above to return the environment in which it was found and its enclosing environment:
-
-
-```r
-fget2 <- function(name, env = caller_env()) {
- # we need only function objects
- f_value <- mget(name,
- envir = env,
- mode = "function",
- inherits = FALSE,
- ifnotfound = list(NULL)
- )
-
- if (!is.null(f_value[[1]])) {
- # success case
- list(
- "where" = env,
- "enclosing" = fn_env(f_value[[1]])
- )
- } else {
- if (!identical(env, empty_env())) {
- # recursive case
- env <- env_parent(env)
- fget2(name, env)
- } else {
- # base case
- stop("No function objects with matching name was found.", call. = FALSE)
- }
- }
-}
-```
-
-Let's try it out:
-
-
-```r
-fget2("mean")
-#> $where
-#>
-
-**Q3.** Write an enhanced version of `str()` that provides more information about functions. Show where the function was found and what environment it was defined in.
-
-**A3.** To write the required function, we can first re-purpose the `fget()` function we wrote above to return the environment in which it was found and its enclosing environment:
-
-
-```r
-fget2 <- function(name, env = caller_env()) {
- # we need only function objects
- f_value <- mget(name,
- envir = env,
- mode = "function",
- inherits = FALSE,
- ifnotfound = list(NULL)
- )
-
- if (!is.null(f_value[[1]])) {
- # success case
- list(
- "where" = env,
- "enclosing" = fn_env(f_value[[1]])
- )
- } else {
- if (!identical(env, empty_env())) {
- # recursive case
- env <- env_parent(env)
- fget2(name, env)
- } else {
- # base case
- stop("No function objects with matching name was found.", call. = FALSE)
- }
- }
-}
-```
-
-Let's try it out:
-
-
-```r
-fget2("mean")
-#> $where
-#>  -
-- `ecdf()`
-
-This function factory computes an empirical cumulative distribution function.
-
-
-```r
-x <- rnorm(12)
-f <- ecdf(x)
-f
-#> Empirical CDF
-#> Call: ecdf(x)
-#> x[1:12] = -1.8793, -1.3221, -1.2392, ..., 1.1604, 1.7956
-f(seq(-2, 2, by = 0.1))
-#> [1] 0.00000000 0.00000000 0.08333333 0.08333333 0.08333333
-#> [6] 0.08333333 0.08333333 0.16666667 0.25000000 0.25000000
-#> [11] 0.33333333 0.33333333 0.33333333 0.41666667 0.41666667
-#> [16] 0.41666667 0.41666667 0.50000000 0.58333333 0.58333333
-#> [21] 0.66666667 0.75000000 0.75000000 0.75000000 0.75000000
-#> [26] 0.75000000 0.75000000 0.75000000 0.75000000 0.83333333
-#> [31] 0.83333333 0.83333333 0.91666667 0.91666667 0.91666667
-#> [36] 0.91666667 0.91666667 0.91666667 1.00000000 1.00000000
-#> [41] 1.00000000
-```
-
----
-
-**Q3.** Create a function `pick()` that takes an index, `i`, as an argument and returns a function with an argument `x` that subsets `x` with `i`.
-
-
-```r
-pick(1)(x)
-# should be equivalent to
-x[[1]]
-
-lapply(mtcars, pick(5))
-# should be equivalent to
-lapply(mtcars, function(x) x[[5]])
-```
-
-**A3.** To write desired function, we just need to make sure that the argument `i` is eagerly evaluated.
-
-
-```r
-pick <- function(i) {
- force(i)
- function(x) x[[i]]
-}
-```
-
-Testing it with specified test cases:
-
-
-```r
-x <- list("a", "b", "c")
-identical(x[[1]], pick(1)(x))
-#> [1] TRUE
-
-identical(
- lapply(mtcars, pick(5)),
- lapply(mtcars, function(x) x[[5]])
-)
-#> [1] TRUE
-```
-
----
-
-**Q4.** Create a function that creates functions that compute the i^th^ [central moment](http://en.wikipedia.org/wiki/Central_moment) of a numeric vector. You can test it by running the following code:
-
-
-```r
-m1 <- moment(1)
-m2 <- moment(2)
-x <- runif(100)
-stopifnot(all.equal(m1(x), 0))
-stopifnot(all.equal(m2(x), var(x) * 99 / 100))
-```
-
-**A4.** The following function satisfied the specified requirements:
-
-
-```r
-moment <- function(k) {
- force(k)
-
- function(x) (sum((x - mean(x))^k)) / length(x)
-}
-```
-
-Testing it with specified test cases:
-
-
-```r
-m1 <- moment(1)
-m2 <- moment(2)
-x <- runif(100)
-
-stopifnot(all.equal(m1(x), 0))
-stopifnot(all.equal(m2(x), var(x) * 99 / 100))
-```
-
----
-
-**Q5.** What happens if you don't use a closure? Make predictions, then verify with the code below.
-
-
-```r
-i <- 0
-new_counter2 <- function() {
- i <<- i + 1
- i
-}
-```
-
-**A5.** In case closures are not used in this context, the counts are stored in a global variable, which can be modified by other processes or even deleted.
-
-
-```r
-new_counter2()
-#> [1] 1
-
-new_counter2()
-#> [1] 2
-
-new_counter2()
-#> [1] 3
-
-i <- 20
-new_counter2()
-#> [1] 21
-```
-
----
-
-**Q6.** What happens if you use `<-` instead of `<<-`? Make predictions, then verify with the code below.
-
-
-```r
-new_counter3 <- function() {
- i <- 0
- function() {
- i <- i + 1
- i
- }
-}
-```
-
-**A6.** In this case, the function will always return `1`.
-
-
-```r
-new_counter3()
-#> function() {
-#> i <- i + 1
-#> i
-#> }
-#>
-
-- `ecdf()`
-
-This function factory computes an empirical cumulative distribution function.
-
-
-```r
-x <- rnorm(12)
-f <- ecdf(x)
-f
-#> Empirical CDF
-#> Call: ecdf(x)
-#> x[1:12] = -1.8793, -1.3221, -1.2392, ..., 1.1604, 1.7956
-f(seq(-2, 2, by = 0.1))
-#> [1] 0.00000000 0.00000000 0.08333333 0.08333333 0.08333333
-#> [6] 0.08333333 0.08333333 0.16666667 0.25000000 0.25000000
-#> [11] 0.33333333 0.33333333 0.33333333 0.41666667 0.41666667
-#> [16] 0.41666667 0.41666667 0.50000000 0.58333333 0.58333333
-#> [21] 0.66666667 0.75000000 0.75000000 0.75000000 0.75000000
-#> [26] 0.75000000 0.75000000 0.75000000 0.75000000 0.83333333
-#> [31] 0.83333333 0.83333333 0.91666667 0.91666667 0.91666667
-#> [36] 0.91666667 0.91666667 0.91666667 1.00000000 1.00000000
-#> [41] 1.00000000
-```
-
----
-
-**Q3.** Create a function `pick()` that takes an index, `i`, as an argument and returns a function with an argument `x` that subsets `x` with `i`.
-
-
-```r
-pick(1)(x)
-# should be equivalent to
-x[[1]]
-
-lapply(mtcars, pick(5))
-# should be equivalent to
-lapply(mtcars, function(x) x[[5]])
-```
-
-**A3.** To write desired function, we just need to make sure that the argument `i` is eagerly evaluated.
-
-
-```r
-pick <- function(i) {
- force(i)
- function(x) x[[i]]
-}
-```
-
-Testing it with specified test cases:
-
-
-```r
-x <- list("a", "b", "c")
-identical(x[[1]], pick(1)(x))
-#> [1] TRUE
-
-identical(
- lapply(mtcars, pick(5)),
- lapply(mtcars, function(x) x[[5]])
-)
-#> [1] TRUE
-```
-
----
-
-**Q4.** Create a function that creates functions that compute the i^th^ [central moment](http://en.wikipedia.org/wiki/Central_moment) of a numeric vector. You can test it by running the following code:
-
-
-```r
-m1 <- moment(1)
-m2 <- moment(2)
-x <- runif(100)
-stopifnot(all.equal(m1(x), 0))
-stopifnot(all.equal(m2(x), var(x) * 99 / 100))
-```
-
-**A4.** The following function satisfied the specified requirements:
-
-
-```r
-moment <- function(k) {
- force(k)
-
- function(x) (sum((x - mean(x))^k)) / length(x)
-}
-```
-
-Testing it with specified test cases:
-
-
-```r
-m1 <- moment(1)
-m2 <- moment(2)
-x <- runif(100)
-
-stopifnot(all.equal(m1(x), 0))
-stopifnot(all.equal(m2(x), var(x) * 99 / 100))
-```
-
----
-
-**Q5.** What happens if you don't use a closure? Make predictions, then verify with the code below.
-
-
-```r
-i <- 0
-new_counter2 <- function() {
- i <<- i + 1
- i
-}
-```
-
-**A5.** In case closures are not used in this context, the counts are stored in a global variable, which can be modified by other processes or even deleted.
-
-
-```r
-new_counter2()
-#> [1] 1
-
-new_counter2()
-#> [1] 2
-
-new_counter2()
-#> [1] 3
-
-i <- 20
-new_counter2()
-#> [1] 21
-```
-
----
-
-**Q6.** What happens if you use `<-` instead of `<<-`? Make predictions, then verify with the code below.
-
-
-```r
-new_counter3 <- function() {
- i <- 0
- function() {
- i <- i + 1
- i
- }
-}
-```
-
-**A6.** In this case, the function will always return `1`.
-
-
-```r
-new_counter3()
-#> function() {
-#> i <- i + 1
-#> i
-#> }
-#>  -
-Or to display axes labels in the desired format:
-
-
-```r
-library(scales)
-
-ggplot(mtcars, aes(wt, mpg)) +
- geom_point() +
- scale_y_continuous(labels = number_format(accuracy = 0.01, decimal.mark = ","))
-```
-
-
-
-Or to display axes labels in the desired format:
-
-
-```r
-library(scales)
-
-ggplot(mtcars, aes(wt, mpg)) +
- geom_point() +
- scale_y_continuous(labels = number_format(accuracy = 0.01, decimal.mark = ","))
-```
-
- -
-The `ggplot2::label_bquote()` adds an additional class to the returned function.
-
-The `scales::number_format()` function is a simple pass-through method that forces evaluation of all its parameters and passes them on to the underlying `scales::number()` function.
-
----
-
-## Statistical factories (Exercises 10.4.4)
-
----
-
-**Q1.** In `boot_model()`, why don't I need to force the evaluation of `df` or `model`?
-
-**A1.** We don’t need to force the evaluation of `df` or `model` because these arguments are automatically evaluated by `lm()`:
-
-
-```r
-boot_model <- function(df, formula) {
- mod <- lm(formula, data = df)
- fitted <- unname(fitted(mod))
- resid <- unname(resid(mod))
- rm(mod)
-
- function() {
- fitted + sample(resid)
- }
-}
-```
-
----
-
-**Q2.** Why might you formulate the Box-Cox transformation like this?
-
-
-```r
-boxcox3 <- function(x) {
- function(lambda) {
- if (lambda == 0) {
- log(x)
- } else {
- (x^lambda - 1) / lambda
- }
- }
-}
-```
-
-**A2.** To see why we formulate this transformation like above, we can compare it to the one mentioned in the book:
-
-
-```r
-boxcox2 <- function(lambda) {
- if (lambda == 0) {
- function(x) log(x)
- } else {
- function(x) (x^lambda - 1) / lambda
- }
-}
-```
-
-Let's have a look at one example with each:
-
-
-```r
-boxcox2(1)
-#> function(x) (x^lambda - 1) / lambda
-#>
-
-The `ggplot2::label_bquote()` adds an additional class to the returned function.
-
-The `scales::number_format()` function is a simple pass-through method that forces evaluation of all its parameters and passes them on to the underlying `scales::number()` function.
-
----
-
-## Statistical factories (Exercises 10.4.4)
-
----
-
-**Q1.** In `boot_model()`, why don't I need to force the evaluation of `df` or `model`?
-
-**A1.** We don’t need to force the evaluation of `df` or `model` because these arguments are automatically evaluated by `lm()`:
-
-
-```r
-boot_model <- function(df, formula) {
- mod <- lm(formula, data = df)
- fitted <- unname(fitted(mod))
- resid <- unname(resid(mod))
- rm(mod)
-
- function() {
- fitted + sample(resid)
- }
-}
-```
-
----
-
-**Q2.** Why might you formulate the Box-Cox transformation like this?
-
-
-```r
-boxcox3 <- function(x) {
- function(lambda) {
- if (lambda == 0) {
- log(x)
- } else {
- (x^lambda - 1) / lambda
- }
- }
-}
-```
-
-**A2.** To see why we formulate this transformation like above, we can compare it to the one mentioned in the book:
-
-
-```r
-boxcox2 <- function(lambda) {
- if (lambda == 0) {
- function(x) log(x)
- } else {
- function(x) (x^lambda - 1) / lambda
- }
-}
-```
-
-Let's have a look at one example with each:
-
-
-```r
-boxcox2(1)
-#> function(x) (x^lambda - 1) / lambda
-#>  -
----
-
-## Function factories + functionals (Exercises 10.5.1)
-
-**Q1.** Which of the following commands is equivalent to `with(x, f(z))`?
-
- (a) `x$f(x$z)`.
- (b) `f(x$z)`.
- (c) `x$f(z)`.
- (d) `f(z)`.
- (e) It depends.
-
-**A1.** It depends on whether `with()` is used with a data frame or a list.
-
-
-```r
-f <- mean
-z <- 1
-x <- list(f = mean, z = 1)
-
-identical(with(x, f(z)), x$f(x$z))
-#> [1] TRUE
-
-identical(with(x, f(z)), f(x$z))
-#> [1] TRUE
-
-identical(with(x, f(z)), x$f(z))
-#> [1] TRUE
-
-identical(with(x, f(z)), f(z))
-#> [1] TRUE
-```
-
----
-
-**Q2.** Compare and contrast the effects of `env_bind()` vs. `attach()` for the following code.
-
-**A2.** Let's compare and contrast the effects of `env_bind()` vs. `attach()`.
-
-- `attach()` adds `funs` to the search path. Since these functions have the same names as functions in `{base}` package, the attached names mask the ones in the `{base}` package.
-
-
-```r
-funs <- list(
- mean = function(x) mean(x, na.rm = TRUE),
- sum = function(x) sum(x, na.rm = TRUE)
-)
-
-attach(funs)
-#> The following objects are masked from package:base:
-#>
-#> mean, sum
-
-mean
-#> function(x) mean(x, na.rm = TRUE)
-head(search())
-#> [1] ".GlobalEnv" "funs" "package:scales"
-#> [4] "package:ggplot2" "package:rlang" "package:magrittr"
-
-mean <- function(x) stop("Hi!")
-mean
-#> function(x) stop("Hi!")
-head(search())
-#> [1] ".GlobalEnv" "funs" "package:scales"
-#> [4] "package:ggplot2" "package:rlang" "package:magrittr"
-
-detach(funs)
-```
-
-- `env_bind()` adds the functions in `funs` to the global environment, instead of masking the names in the `{base}` package.
-
-
-```r
-env_bind(globalenv(), !!!funs)
-mean
-#> function(x) mean(x, na.rm = TRUE)
-
-mean <- function(x) stop("Hi!")
-mean
-#> function(x) stop("Hi!")
-env_unbind(globalenv(), names(funs))
-```
-
-Note that there is no `"funs"` in this output.
-
----
-
-## Session information
-
-
-```r
-sessioninfo::session_info(include_base = TRUE)
-#> ─ Session info ───────────────────────────────────────────
-#> setting value
-#> version R version 4.2.2 (2022-10-31)
-#> os macOS Ventura 13.0
-#> system aarch64, darwin20
-#> ui X11
-#> language (EN)
-#> collate en_US.UTF-8
-#> ctype en_US.UTF-8
-#> tz Europe/Berlin
-#> date 2022-11-12
-#> pandoc 2.19.2 @ /Applications/RStudio.app/Contents/MacOS/quarto/bin/tools/ (via rmarkdown)
-#>
-#> ─ Packages ───────────────────────────────────────────────
-#> ! package * version date (UTC) lib source
-#> assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.2.0)
-#> base * 4.2.2 2022-10-31 [?] local
-#> bench 1.1.2 2021-11-30 [1] CRAN (R 4.2.0)
-#> bookdown 0.30 2022-11-09 [1] CRAN (R 4.2.2)
-#> bslib 0.4.1 2022-11-02 [1] CRAN (R 4.2.2)
-#> cachem 1.0.6 2021-08-19 [1] CRAN (R 4.2.0)
-#> cli 3.4.1 2022-09-23 [1] CRAN (R 4.2.0)
-#> colorspace 2.0-3 2022-02-21 [1] CRAN (R 4.2.0)
-#> P compiler 4.2.2 2022-10-31 [1] local
-#> P datasets * 4.2.2 2022-10-31 [1] local
-#> DBI 1.1.3.9002 2022-10-17 [1] Github (r-dbi/DBI@2aec388)
-#> digest 0.6.30 2022-10-18 [1] CRAN (R 4.2.1)
-#> downlit 0.4.2 2022-07-05 [1] CRAN (R 4.2.1)
-#> dplyr 1.0.10 2022-09-01 [1] CRAN (R 4.2.1)
-#> evaluate 0.18 2022-11-07 [1] CRAN (R 4.2.2)
-#> fansi 1.0.3 2022-03-24 [1] CRAN (R 4.2.0)
-#> farver 2.1.1 2022-07-06 [1] CRAN (R 4.2.1)
-#> fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.2.0)
-#> fs 1.5.2 2021-12-08 [1] CRAN (R 4.2.0)
-#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.2.1)
-#> ggplot2 * 3.4.0 2022-11-04 [1] CRAN (R 4.2.2)
-#> glue 1.6.2 2022-02-24 [1] CRAN (R 4.2.0)
-#> P graphics * 4.2.2 2022-10-31 [1] local
-#> P grDevices * 4.2.2 2022-10-31 [1] local
-#> P grid 4.2.2 2022-10-31 [1] local
-#> gtable 0.3.1 2022-09-01 [1] CRAN (R 4.2.1)
-#> highr 0.9 2021-04-16 [1] CRAN (R 4.2.0)
-#> htmltools 0.5.3 2022-07-18 [1] CRAN (R 4.2.1)
-#> jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.2.0)
-#> jsonlite 1.8.3 2022-10-21 [1] CRAN (R 4.2.1)
-#> knitr 1.40 2022-08-24 [1] CRAN (R 4.2.1)
-#> labeling 0.4.2 2020-10-20 [1] CRAN (R 4.2.0)
-#> lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.2.1)
-#> lobstr 1.1.2 2022-06-22 [1] CRAN (R 4.2.0)
-#> magrittr * 2.0.3 2022-03-30 [1] CRAN (R 4.2.0)
-#> memoise 2.0.1 2021-11-26 [1] CRAN (R 4.2.0)
-#> P methods * 4.2.2 2022-10-31 [1] local
-#> munsell 0.5.0 2018-06-12 [1] CRAN (R 4.2.0)
-#> pillar 1.8.1 2022-08-19 [1] CRAN (R 4.2.1)
-#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.2.0)
-#> profmem 0.6.0 2020-12-13 [1] CRAN (R 4.2.0)
-#> purrr 0.3.5 2022-10-06 [1] CRAN (R 4.2.1)
-#> R6 2.5.1.9000 2022-10-27 [1] local
-#> rlang * 1.0.6 2022-09-24 [1] CRAN (R 4.2.1)
-#> rmarkdown 2.18 2022-11-09 [1] CRAN (R 4.2.2)
-#> rstudioapi 0.14 2022-08-22 [1] CRAN (R 4.2.1)
-#> sass 0.4.2 2022-07-16 [1] CRAN (R 4.2.1)
-#> scales * 1.2.1 2022-08-20 [1] CRAN (R 4.2.1)
-#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.2.0)
-#> P stats * 4.2.2 2022-10-31 [1] local
-#> stringi 1.7.8 2022-07-11 [1] CRAN (R 4.2.1)
-#> stringr 1.4.1 2022-08-20 [1] CRAN (R 4.2.1)
-#> tibble 3.1.8.9002 2022-10-16 [1] local
-#> tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.2.1)
-#> P tools 4.2.2 2022-10-31 [1] local
-#> utf8 1.2.2 2021-07-24 [1] CRAN (R 4.2.0)
-#> P utils * 4.2.2 2022-10-31 [1] local
-#> vctrs 0.5.0 2022-10-22 [1] CRAN (R 4.2.1)
-#> withr 2.5.0 2022-03-03 [1] CRAN (R 4.2.0)
-#> xfun 0.34 2022-10-18 [1] CRAN (R 4.2.1)
-#> xml2 1.3.3.9000 2022-10-10 [1] local
-#> yaml 2.3.6 2022-10-18 [1] CRAN (R 4.2.1)
-#>
-#> [1] /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library
-#>
-#> P ── Loaded and on-disk path mismatch.
-#>
-#> ──────────────────────────────────────────────────────────
-```
-
diff --git a/_book/Function-factories_files/figure-html/Function-factories-18-1.png b/_book/Function-factories_files/figure-html/Function-factories-18-1.png
deleted file mode 100644
index 1e696e5e..00000000
Binary files a/_book/Function-factories_files/figure-html/Function-factories-18-1.png and /dev/null differ
diff --git a/_book/Function-factories_files/figure-html/Function-factories-19-1.png b/_book/Function-factories_files/figure-html/Function-factories-19-1.png
deleted file mode 100644
index 9cb4b9e3..00000000
Binary files a/_book/Function-factories_files/figure-html/Function-factories-19-1.png and /dev/null differ
diff --git a/_book/Function-factories_files/figure-html/Function-factories-28-1.png b/_book/Function-factories_files/figure-html/Function-factories-28-1.png
deleted file mode 100644
index 04c05abc..00000000
Binary files a/_book/Function-factories_files/figure-html/Function-factories-28-1.png and /dev/null differ
diff --git a/_book/Function-factories_files/figure-html/Function-factories-4-1.png b/_book/Function-factories_files/figure-html/Function-factories-4-1.png
deleted file mode 100644
index 0c37642d..00000000
Binary files a/_book/Function-factories_files/figure-html/Function-factories-4-1.png and /dev/null differ
diff --git a/_book/Function-operators.md b/_book/Function-operators.md
deleted file mode 100644
index e685bfd9..00000000
--- a/_book/Function-operators.md
+++ /dev/null
@@ -1,398 +0,0 @@
-# Function operators
-
-
-
-Attaching the needed libraries:
-
-
-```r
-library(purrr, warn.conflicts = FALSE)
-```
-
-## Existing function operators (Exercises 11.2.3)
-
----
-
-**Q1.** Base R provides a function operator in the form of `Vectorize()`. What does it do? When might you use it?
-
-**A1.** `Vectorize()` function creates a function that vectorizes the action of the provided function over specified arguments (i.e., it acts on each element of the vector). We will see its utility by solving a problem that otherwise would be difficult to solve.
-
-The problem is to find indices of matching numeric values for the given threshold by creating a hybrid of the following functions:
-
-- `%in%` (which doesn't provide any way to provide tolerance when comparing numeric values),
-- `dplyr::near()` (which is vectorized element-wise and thus expects two vectors of equal length)
-
-
-```r
-which_near <- function(x, y, tolerance) {
- # Vectorize `dplyr::near()` function only over the `y` argument.
- # `Vectorize()` is a function operator and will return a function.
- customNear <- Vectorize(dplyr::near, vectorize.args = c("y"), SIMPLIFY = FALSE)
-
- # Apply the vectorized function to vector arguments and then check where the
- # comparisons are equal (i.e. `TRUE`) using `which()`.
- #
- # Use `compact()` to remove empty elements from the resulting list.
- index_list <- purrr::compact(purrr::map(customNear(x, y, tol = tolerance), which))
-
- # If there are any matches, return the indices as an atomic vector of integers.
- if (length(index_list) > 0L) {
- index_vector <- purrr::simplify(index_list, "integer")
- return(index_vector)
- }
-
- # If there are no matches
- return(integer(0L))
-}
-```
-
-Let's use it:
-
-
-```r
-x1 <- c(2.1, 3.3, 8.45, 8, 6)
-x2 <- c(6, 8.40, 3)
-
-which_near(x1, x2, tolerance = 0.1)
-#> [1] 5 3
-```
-
-Note that we needed to create a new function for this because neither of the existing functions do what we want.
-
-
-```r
-which(x1 %in% x2)
-#> [1] 5
-
-which(dplyr::near(x1, x2, tol = 0.1))
-#> Warning in x - y: longer object length is not a multiple of
-#> shorter object length
-#> integer(0)
-```
-
-We solved a complex task here using the `Vectorize()` function!
-
----
-
-**Q2.** Read the source code for `possibly()`. How does it work?
-
-**A2.** Let's have a look at the source code for this function:
-
-
-```r
-possibly
-#> function (.f, otherwise, quiet = TRUE)
-#> {
-#> .f <- as_mapper(.f)
-#> force(otherwise)
-#> function(...) {
-#> tryCatch(.f(...), error = function(e) {
-#> if (!quiet)
-#> message("Error: ", e$message)
-#> otherwise
-#> }, interrupt = function(e) {
-#> stop("Terminated by user", call. = FALSE)
-#> })
-#> }
-#> }
-#>
-
----
-
-## Function factories + functionals (Exercises 10.5.1)
-
-**Q1.** Which of the following commands is equivalent to `with(x, f(z))`?
-
- (a) `x$f(x$z)`.
- (b) `f(x$z)`.
- (c) `x$f(z)`.
- (d) `f(z)`.
- (e) It depends.
-
-**A1.** It depends on whether `with()` is used with a data frame or a list.
-
-
-```r
-f <- mean
-z <- 1
-x <- list(f = mean, z = 1)
-
-identical(with(x, f(z)), x$f(x$z))
-#> [1] TRUE
-
-identical(with(x, f(z)), f(x$z))
-#> [1] TRUE
-
-identical(with(x, f(z)), x$f(z))
-#> [1] TRUE
-
-identical(with(x, f(z)), f(z))
-#> [1] TRUE
-```
-
----
-
-**Q2.** Compare and contrast the effects of `env_bind()` vs. `attach()` for the following code.
-
-**A2.** Let's compare and contrast the effects of `env_bind()` vs. `attach()`.
-
-- `attach()` adds `funs` to the search path. Since these functions have the same names as functions in `{base}` package, the attached names mask the ones in the `{base}` package.
-
-
-```r
-funs <- list(
- mean = function(x) mean(x, na.rm = TRUE),
- sum = function(x) sum(x, na.rm = TRUE)
-)
-
-attach(funs)
-#> The following objects are masked from package:base:
-#>
-#> mean, sum
-
-mean
-#> function(x) mean(x, na.rm = TRUE)
-head(search())
-#> [1] ".GlobalEnv" "funs" "package:scales"
-#> [4] "package:ggplot2" "package:rlang" "package:magrittr"
-
-mean <- function(x) stop("Hi!")
-mean
-#> function(x) stop("Hi!")
-head(search())
-#> [1] ".GlobalEnv" "funs" "package:scales"
-#> [4] "package:ggplot2" "package:rlang" "package:magrittr"
-
-detach(funs)
-```
-
-- `env_bind()` adds the functions in `funs` to the global environment, instead of masking the names in the `{base}` package.
-
-
-```r
-env_bind(globalenv(), !!!funs)
-mean
-#> function(x) mean(x, na.rm = TRUE)
-
-mean <- function(x) stop("Hi!")
-mean
-#> function(x) stop("Hi!")
-env_unbind(globalenv(), names(funs))
-```
-
-Note that there is no `"funs"` in this output.
-
----
-
-## Session information
-
-
-```r
-sessioninfo::session_info(include_base = TRUE)
-#> ─ Session info ───────────────────────────────────────────
-#> setting value
-#> version R version 4.2.2 (2022-10-31)
-#> os macOS Ventura 13.0
-#> system aarch64, darwin20
-#> ui X11
-#> language (EN)
-#> collate en_US.UTF-8
-#> ctype en_US.UTF-8
-#> tz Europe/Berlin
-#> date 2022-11-12
-#> pandoc 2.19.2 @ /Applications/RStudio.app/Contents/MacOS/quarto/bin/tools/ (via rmarkdown)
-#>
-#> ─ Packages ───────────────────────────────────────────────
-#> ! package * version date (UTC) lib source
-#> assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.2.0)
-#> base * 4.2.2 2022-10-31 [?] local
-#> bench 1.1.2 2021-11-30 [1] CRAN (R 4.2.0)
-#> bookdown 0.30 2022-11-09 [1] CRAN (R 4.2.2)
-#> bslib 0.4.1 2022-11-02 [1] CRAN (R 4.2.2)
-#> cachem 1.0.6 2021-08-19 [1] CRAN (R 4.2.0)
-#> cli 3.4.1 2022-09-23 [1] CRAN (R 4.2.0)
-#> colorspace 2.0-3 2022-02-21 [1] CRAN (R 4.2.0)
-#> P compiler 4.2.2 2022-10-31 [1] local
-#> P datasets * 4.2.2 2022-10-31 [1] local
-#> DBI 1.1.3.9002 2022-10-17 [1] Github (r-dbi/DBI@2aec388)
-#> digest 0.6.30 2022-10-18 [1] CRAN (R 4.2.1)
-#> downlit 0.4.2 2022-07-05 [1] CRAN (R 4.2.1)
-#> dplyr 1.0.10 2022-09-01 [1] CRAN (R 4.2.1)
-#> evaluate 0.18 2022-11-07 [1] CRAN (R 4.2.2)
-#> fansi 1.0.3 2022-03-24 [1] CRAN (R 4.2.0)
-#> farver 2.1.1 2022-07-06 [1] CRAN (R 4.2.1)
-#> fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.2.0)
-#> fs 1.5.2 2021-12-08 [1] CRAN (R 4.2.0)
-#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.2.1)
-#> ggplot2 * 3.4.0 2022-11-04 [1] CRAN (R 4.2.2)
-#> glue 1.6.2 2022-02-24 [1] CRAN (R 4.2.0)
-#> P graphics * 4.2.2 2022-10-31 [1] local
-#> P grDevices * 4.2.2 2022-10-31 [1] local
-#> P grid 4.2.2 2022-10-31 [1] local
-#> gtable 0.3.1 2022-09-01 [1] CRAN (R 4.2.1)
-#> highr 0.9 2021-04-16 [1] CRAN (R 4.2.0)
-#> htmltools 0.5.3 2022-07-18 [1] CRAN (R 4.2.1)
-#> jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.2.0)
-#> jsonlite 1.8.3 2022-10-21 [1] CRAN (R 4.2.1)
-#> knitr 1.40 2022-08-24 [1] CRAN (R 4.2.1)
-#> labeling 0.4.2 2020-10-20 [1] CRAN (R 4.2.0)
-#> lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.2.1)
-#> lobstr 1.1.2 2022-06-22 [1] CRAN (R 4.2.0)
-#> magrittr * 2.0.3 2022-03-30 [1] CRAN (R 4.2.0)
-#> memoise 2.0.1 2021-11-26 [1] CRAN (R 4.2.0)
-#> P methods * 4.2.2 2022-10-31 [1] local
-#> munsell 0.5.0 2018-06-12 [1] CRAN (R 4.2.0)
-#> pillar 1.8.1 2022-08-19 [1] CRAN (R 4.2.1)
-#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.2.0)
-#> profmem 0.6.0 2020-12-13 [1] CRAN (R 4.2.0)
-#> purrr 0.3.5 2022-10-06 [1] CRAN (R 4.2.1)
-#> R6 2.5.1.9000 2022-10-27 [1] local
-#> rlang * 1.0.6 2022-09-24 [1] CRAN (R 4.2.1)
-#> rmarkdown 2.18 2022-11-09 [1] CRAN (R 4.2.2)
-#> rstudioapi 0.14 2022-08-22 [1] CRAN (R 4.2.1)
-#> sass 0.4.2 2022-07-16 [1] CRAN (R 4.2.1)
-#> scales * 1.2.1 2022-08-20 [1] CRAN (R 4.2.1)
-#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.2.0)
-#> P stats * 4.2.2 2022-10-31 [1] local
-#> stringi 1.7.8 2022-07-11 [1] CRAN (R 4.2.1)
-#> stringr 1.4.1 2022-08-20 [1] CRAN (R 4.2.1)
-#> tibble 3.1.8.9002 2022-10-16 [1] local
-#> tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.2.1)
-#> P tools 4.2.2 2022-10-31 [1] local
-#> utf8 1.2.2 2021-07-24 [1] CRAN (R 4.2.0)
-#> P utils * 4.2.2 2022-10-31 [1] local
-#> vctrs 0.5.0 2022-10-22 [1] CRAN (R 4.2.1)
-#> withr 2.5.0 2022-03-03 [1] CRAN (R 4.2.0)
-#> xfun 0.34 2022-10-18 [1] CRAN (R 4.2.1)
-#> xml2 1.3.3.9000 2022-10-10 [1] local
-#> yaml 2.3.6 2022-10-18 [1] CRAN (R 4.2.1)
-#>
-#> [1] /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library
-#>
-#> P ── Loaded and on-disk path mismatch.
-#>
-#> ──────────────────────────────────────────────────────────
-```
-
diff --git a/_book/Function-factories_files/figure-html/Function-factories-18-1.png b/_book/Function-factories_files/figure-html/Function-factories-18-1.png
deleted file mode 100644
index 1e696e5e..00000000
Binary files a/_book/Function-factories_files/figure-html/Function-factories-18-1.png and /dev/null differ
diff --git a/_book/Function-factories_files/figure-html/Function-factories-19-1.png b/_book/Function-factories_files/figure-html/Function-factories-19-1.png
deleted file mode 100644
index 9cb4b9e3..00000000
Binary files a/_book/Function-factories_files/figure-html/Function-factories-19-1.png and /dev/null differ
diff --git a/_book/Function-factories_files/figure-html/Function-factories-28-1.png b/_book/Function-factories_files/figure-html/Function-factories-28-1.png
deleted file mode 100644
index 04c05abc..00000000
Binary files a/_book/Function-factories_files/figure-html/Function-factories-28-1.png and /dev/null differ
diff --git a/_book/Function-factories_files/figure-html/Function-factories-4-1.png b/_book/Function-factories_files/figure-html/Function-factories-4-1.png
deleted file mode 100644
index 0c37642d..00000000
Binary files a/_book/Function-factories_files/figure-html/Function-factories-4-1.png and /dev/null differ
diff --git a/_book/Function-operators.md b/_book/Function-operators.md
deleted file mode 100644
index e685bfd9..00000000
--- a/_book/Function-operators.md
+++ /dev/null
@@ -1,398 +0,0 @@
-# Function operators
-
-
-
-Attaching the needed libraries:
-
-
-```r
-library(purrr, warn.conflicts = FALSE)
-```
-
-## Existing function operators (Exercises 11.2.3)
-
----
-
-**Q1.** Base R provides a function operator in the form of `Vectorize()`. What does it do? When might you use it?
-
-**A1.** `Vectorize()` function creates a function that vectorizes the action of the provided function over specified arguments (i.e., it acts on each element of the vector). We will see its utility by solving a problem that otherwise would be difficult to solve.
-
-The problem is to find indices of matching numeric values for the given threshold by creating a hybrid of the following functions:
-
-- `%in%` (which doesn't provide any way to provide tolerance when comparing numeric values),
-- `dplyr::near()` (which is vectorized element-wise and thus expects two vectors of equal length)
-
-
-```r
-which_near <- function(x, y, tolerance) {
- # Vectorize `dplyr::near()` function only over the `y` argument.
- # `Vectorize()` is a function operator and will return a function.
- customNear <- Vectorize(dplyr::near, vectorize.args = c("y"), SIMPLIFY = FALSE)
-
- # Apply the vectorized function to vector arguments and then check where the
- # comparisons are equal (i.e. `TRUE`) using `which()`.
- #
- # Use `compact()` to remove empty elements from the resulting list.
- index_list <- purrr::compact(purrr::map(customNear(x, y, tol = tolerance), which))
-
- # If there are any matches, return the indices as an atomic vector of integers.
- if (length(index_list) > 0L) {
- index_vector <- purrr::simplify(index_list, "integer")
- return(index_vector)
- }
-
- # If there are no matches
- return(integer(0L))
-}
-```
-
-Let's use it:
-
-
-```r
-x1 <- c(2.1, 3.3, 8.45, 8, 6)
-x2 <- c(6, 8.40, 3)
-
-which_near(x1, x2, tolerance = 0.1)
-#> [1] 5 3
-```
-
-Note that we needed to create a new function for this because neither of the existing functions do what we want.
-
-
-```r
-which(x1 %in% x2)
-#> [1] 5
-
-which(dplyr::near(x1, x2, tol = 0.1))
-#> Warning in x - y: longer object length is not a multiple of
-#> shorter object length
-#> integer(0)
-```
-
-We solved a complex task here using the `Vectorize()` function!
-
----
-
-**Q2.** Read the source code for `possibly()`. How does it work?
-
-**A2.** Let's have a look at the source code for this function:
-
-
-```r
-possibly
-#> function (.f, otherwise, quiet = TRUE)
-#> {
-#> .f <- as_mapper(.f)
-#> force(otherwise)
-#> function(...) {
-#> tryCatch(.f(...), error = function(e) {
-#> if (!quiet)
-#> message("Error: ", e$message)
-#> otherwise
-#> }, interrupt = function(e) {
-#> stop("Terminated by user", call. = FALSE)
-#> })
-#> }
-#> }
-#>  -
-```r
-
-hist(p)
-```
-
-
-
-```r
-
-hist(p)
-```
-
- -
----
-
-**Q5.** The following code uses a map nested inside another map to apply a function to every element of a nested list. Why does it fail, and what do you need to do to make it work?
-
-
-```r
-x <- list(
- list(1, c(3, 9)),
- list(c(3, 6), 7, c(4, 7, 6))
-)
-
-triple <- function(x) x * 3
-map(x, map, .f = triple)
-#> Error in .f(.x[[i]], ...): unused argument (function (.x, .f, ...)
-#> {
-#> .f <- as_mapper(.f, ...)
-#> .Call(map_impl, environment(), ".x", ".f", "list")
-#> })
-```
-
-**A5.** This function fails because this call effectively evaluates to the following:
-
-
-```r
-map(.x = x, .f = ~ triple(x = .x, map))
-```
-
-But `triple()` has only one parameter (`x`), and so the execution fails.
-
-Here is the fixed version:
-
-
-```r
-x <- list(
- list(1, c(3, 9)),
- list(c(3, 6), 7, c(4, 7, 6))
-)
-
-triple <- function(x) x * 3
-map(x, .f = ~ map(.x, ~ triple(.x)))
-#> [[1]]
-#> [[1]][[1]]
-#> [1] 3
-#>
-#> [[1]][[2]]
-#> [1] 9 27
-#>
-#>
-#> [[2]]
-#> [[2]][[1]]
-#> [1] 9 18
-#>
-#> [[2]][[2]]
-#> [1] 21
-#>
-#> [[2]][[3]]
-#> [1] 12 21 18
-```
-
----
-
-**Q6.** Use `map()` to fit linear models to the `mtcars` dataset using the formulas stored in this list:
-
-
-```r
-formulas <- list(
- mpg ~ disp,
- mpg ~ I(1 / disp),
- mpg ~ disp + wt,
- mpg ~ I(1 / disp) + wt
-)
-```
-
-**A6.** Fitting linear models to the `mtcars` dataset using the provided formulas:
-
-
-```r
-formulas <- list(
- mpg ~ disp,
- mpg ~ I(1 / disp),
- mpg ~ disp + wt,
- mpg ~ I(1 / disp) + wt
-)
-
-map(formulas, ~ lm(formula = ., data = mtcars))
-#> [[1]]
-#>
-#> Call:
-#> lm(formula = ., data = mtcars)
-#>
-#> Coefficients:
-#> (Intercept) disp
-#> 29.59985 -0.04122
-#>
-#>
-#> [[2]]
-#>
-#> Call:
-#> lm(formula = ., data = mtcars)
-#>
-#> Coefficients:
-#> (Intercept) I(1/disp)
-#> 10.75 1557.67
-#>
-#>
-#> [[3]]
-#>
-#> Call:
-#> lm(formula = ., data = mtcars)
-#>
-#> Coefficients:
-#> (Intercept) disp wt
-#> 34.96055 -0.01772 -3.35083
-#>
-#>
-#> [[4]]
-#>
-#> Call:
-#> lm(formula = ., data = mtcars)
-#>
-#> Coefficients:
-#> (Intercept) I(1/disp) wt
-#> 19.024 1142.560 -1.798
-```
-
----
-
-**Q7.** Fit the model `mpg ~ disp` to each of the bootstrap replicates of `mtcars` in the list below, then extract the $R^2$ of the model fit (Hint: you can compute the $R^2$ with `summary()`.)
-
-
-```r
-bootstrap <- function(df) {
- df[sample(nrow(df), replace = TRUE), , drop = FALSE]
-}
-
-bootstraps <- map(1:10, ~ bootstrap(mtcars))
-```
-
-**A7.** This can be done using `map_dbl()`:
-
-
-```r
-bootstrap <- function(df) {
- df[sample(nrow(df), replace = TRUE), , drop = FALSE]
-}
-
-bootstraps <- map(1:10, ~ bootstrap(mtcars))
-
-bootstraps %>%
- map(~ lm(mpg ~ disp, data = .x)) %>%
- map(summary) %>%
- map_dbl("r.squared")
-#> [1] 0.7864562 0.8110818 0.7956331 0.7632399 0.7967824
-#> [6] 0.7364226 0.7203027 0.6653252 0.7732780 0.6753329
-```
-
----
-
-## Map variants (Exercises 9.4.6)
-
----
-
-**Q1.** Explain the results of `modify(mtcars, 1)`.
-
-**A1.** `modify()` returns the object of type same as the input. Since the input here is a data frame of certain dimensions and `.f = 1` translates to plucking the first element in each column, it returns a data frame with the same dimensions with the plucked element recycled across rows.
-
-
-```r
-head(modify(mtcars, 1))
-#> mpg cyl disp hp drat wt qsec vs am
-#> Mazda RX4 21 6 160 110 3.9 2.62 16.46 0 1
-#> Mazda RX4 Wag 21 6 160 110 3.9 2.62 16.46 0 1
-#> Datsun 710 21 6 160 110 3.9 2.62 16.46 0 1
-#> Hornet 4 Drive 21 6 160 110 3.9 2.62 16.46 0 1
-#> Hornet Sportabout 21 6 160 110 3.9 2.62 16.46 0 1
-#> Valiant 21 6 160 110 3.9 2.62 16.46 0 1
-#> gear carb
-#> Mazda RX4 4 4
-#> Mazda RX4 Wag 4 4
-#> Datsun 710 4 4
-#> Hornet 4 Drive 4 4
-#> Hornet Sportabout 4 4
-#> Valiant 4 4
-```
-
----
-
-**Q2.** Rewrite the following code to use `iwalk()` instead of `walk2()`. What are the advantages and disadvantages?
-
-
-```r
-cyls <- split(mtcars, mtcars$cyl)
-paths <- file.path(temp, paste0("cyl-", names(cyls), ".csv"))
-walk2(cyls, paths, write.csv)
-```
-
-**A2.** Let's first rewrite provided code using `iwalk()`:
-
-
-```r
-cyls <- split(mtcars, mtcars$cyl)
-names(cyls) <- file.path(temp, paste0("cyl-", names(cyls), ".csv"))
-iwalk(cyls, ~ write.csv(.x, .y))
-```
-
-The advantage of using `iwalk()` is that we need to now deal with only a single variable (`cyls`) instead of two (`cyls` and `paths`).
-
-The disadvantage is that the code is difficult to reason about:
-In `walk2()`, it's explicit what `.x` (`= cyls`) and `.y` (`= paths`) correspond to, while this is not so for `iwalk()` (i.e., `.x = cyls` and `.y = names(cyls)`) with the `.y` argument being "invisible".
-
----
-
-**Q3.** Explain how the following code transforms a data frame using functions stored in a list.
-
-
-```r
-trans <- list(
- disp = function(x) x * 0.0163871,
- am = function(x) factor(x, labels = c("auto", "manual"))
-)
-
-nm <- names(trans)
-mtcars[nm] <- map2(trans, mtcars[nm], function(f, var) f(var))
-```
-
-Compare and contrast the `map2()` approach to this `map()` approach:
-
-
-```r
-mtcars[nm] <- map(nm, ~ trans[[.x]](mtcars[[.x]]))
-```
-
-**A3.** `map2()` supplies the functions stored in `trans` as anonymous functions via placeholder `f`, while the names of the columns specified in `mtcars[nm]` are supplied as `var` argument to the anonymous function. Note that the function is iterating over indices for vectors of transformations and column names.
-
-
-```r
-trans <- list(
- disp = function(x) x * 0.0163871,
- am = function(x) factor(x, labels = c("auto", "manual"))
-)
-
-nm <- names(trans)
-mtcars[nm] <- map2(trans, mtcars[nm], function(f, var) f(var))
-```
-
-In the `map()` approach, the function is iterating over indices for vectors of column names.
-
-
-```r
-mtcars[nm] <- map(nm, ~ trans[[.x]](mtcars[[.x]]))
-```
-
-The latter approach can't afford passing arguments to placeholders in an anonymous function.
-
----
-
-**Q4.** What does `write.csv()` return, i.e. what happens if you use it with `map2()` instead of `walk2()`?
-
-**A4.** If we use `map2()`, it will work, but it will print `NULL`s to the console for every list element.
-
-
-```r
-withr::with_tempdir(
- code = {
- ls <- split(mtcars, mtcars$cyl)
- nm <- names(ls)
- map2(ls, nm, write.csv)
- }
-)
-#> $`4`
-#> NULL
-#>
-#> $`6`
-#> NULL
-#>
-#> $`8`
-#> NULL
-```
-
----
-
-## Predicate functionals (Exercises 9.6.3)
-
----
-
-**Q1.** Why isn't `is.na()` a predicate function? What base R function is closest to being a predicate version of `is.na()`?
-
-**A1.** As mentioned in the docs:
-
-> A predicate is a function that returns a **single** `TRUE` or `FALSE`.
-
-The `is.na()` function does not return a `logical` scalar, but instead returns a vector and thus isn't a predicate function.
-
-
-```r
-# contrast the following behavior of predicate functions
-is.character(c("x", 2))
-#> [1] TRUE
-is.null(c(3, NULL))
-#> [1] FALSE
-
-# with this behavior
-is.na(c(NA, 1))
-#> [1] TRUE FALSE
-```
-
-The closest equivalent of a predicate function in base-R is `anyNA()` function.
-
-
-```r
-anyNA(c(NA, 1))
-#> [1] TRUE
-```
-
----
-
-**Q2.** `simple_reduce()` has a problem when `x` is length 0 or length 1. Describe the source of the problem and how you might go about fixing it.
-
-
-```r
-simple_reduce <- function(x, f) {
- out <- x[[1]]
- for (i in seq(2, length(x))) {
- out <- f(out, x[[i]])
- }
- out
-}
-```
-
-**A2.** The supplied function struggles with inputs of length 0 and 1 because function tries to subscript out-of-bound values.
-
-
-```r
-simple_reduce(numeric(), sum)
-#> Error in x[[1]]: subscript out of bounds
-simple_reduce(1, sum)
-#> Error in x[[i]]: subscript out of bounds
-simple_reduce(1:3, sum)
-#> [1] 6
-```
-
-This problem can be solved by adding `init` argument, which supplies the default or initial value:
-
-
-```r
-simple_reduce2 <- function(x, f, init = 0) {
- # initializer will become the first value
- if (length(x) == 0L) {
- return(init)
- }
-
- if (length(x) == 1L) {
- return(x[[1L]])
- }
-
- out <- x[[1]]
-
- for (i in seq(2, length(x))) {
- out <- f(out, x[[i]])
- }
-
- out
-}
-```
-
-Let's try it out:
-
-
-```r
-simple_reduce2(numeric(), sum)
-#> [1] 0
-simple_reduce2(1, sum)
-#> [1] 1
-simple_reduce2(1:3, sum)
-#> [1] 6
-```
-
-Depending on the function, we can provide a different `init` argument:
-
-
-```r
-simple_reduce2(numeric(), `*`, init = 1)
-#> [1] 1
-simple_reduce2(1, `*`, init = 1)
-#> [1] 1
-simple_reduce2(1:3, `*`, init = 1)
-#> [1] 6
-```
-
----
-
-**Q3.** Implement the `span()` function from Haskell: given a list `x` and a predicate function `f`, `span(x, f)` returns the location of the longest sequential run of elements where the predicate is true. (Hint: you might find `rle()` helpful.)
-
-**A3.** Implementation of `span()`:
-
-
-```r
-span <- function(x, f) {
- running_lengths <- purrr::map_lgl(x, ~ f(.x)) %>% rle()
-
- df <- dplyr::tibble(
- "lengths" = running_lengths$lengths,
- "values" = running_lengths$values
- ) %>%
- dplyr::mutate(rowid = dplyr::row_number()) %>%
- dplyr::filter(values)
-
- # no sequence where condition is `TRUE`
- if (nrow(df) == 0L) {
- return(integer())
- }
-
- # only single sequence where condition is `TRUE`
- if (nrow(df) == 1L) {
- return((df$rowid):(df$lengths - 1 + df$rowid))
- }
-
- # multiple sequences where condition is `TRUE`; select max one
- if (nrow(df) > 1L) {
- df <- dplyr::filter(df, lengths == max(lengths))
- return((df$rowid):(df$lengths - 1 + df$rowid))
- }
-}
-```
-
-Testing it once:
-
-
-```r
-span(c(0, 0, 0, 0, 0), is.na)
-#> integer(0)
-span(c(NA, 0, NA, NA, NA), is.na)
-#> [1] 3 4 5
-span(c(NA, 0, 0, 0, 0), is.na)
-#> [1] 1
-span(c(NA, NA, 0, 0, 0), is.na)
-#> [1] 1 2
-```
-
-Testing it twice:
-
-
-```r
-span(c(3, 1, 2, 4, 5, 6), function(x) x > 3)
-#> [1] 2 3 4
-span(c(3, 1, 2, 4, 5, 6), function(x) x > 9)
-#> integer(0)
-span(c(3, 1, 2, 4, 5, 6), function(x) x == 3)
-#> [1] 1
-span(c(3, 1, 2, 4, 5, 6), function(x) x %in% c(2, 4))
-#> [1] 2 3
-```
-
----
-
-**Q4.** Implement `arg_max()`. It should take a function and a vector of inputs, and return the elements of the input where the function returns the highest value. For example, `arg_max(-10:5, function(x) x ^ 2)` should return -10. `arg_max(-5:5, function(x) x ^ 2)` should return `c(-5, 5)`. Also implement the matching `arg_min()` function.
-
-**A4.** Here are implementations for the specified functions:
-
-- Implementing `arg_max()`
-
-
-```r
-arg_max <- function(.x, .f) {
- df <- dplyr::tibble(
- original = .x,
- transformed = purrr::map_dbl(.x, .f)
- )
-
- dplyr::filter(df, transformed == max(transformed))[["original"]]
-}
-
-arg_max(-10:5, function(x) x^2)
-#> [1] -10
-arg_max(-5:5, function(x) x^2)
-#> [1] -5 5
-```
-
-- Implementing `arg_min()`
-
-
-```r
-arg_min <- function(.x, .f) {
- df <- dplyr::tibble(
- original = .x,
- transformed = purrr::map_dbl(.x, .f)
- )
-
- dplyr::filter(df, transformed == min(transformed))[["original"]]
-}
-
-arg_min(-10:5, function(x) x^2)
-#> [1] 0
-arg_min(-5:5, function(x) x^2)
-#> [1] 0
-```
-
----
-
-**Q5.** The function below scales a vector so it falls in the range [0, 1]. How would you apply it to every column of a data frame? How would you apply it to every numeric column in a data frame?
-
-
-```r
-scale01 <- function(x) {
- rng <- range(x, na.rm = TRUE)
- (x - rng[1]) / (rng[2] - rng[1])
-}
-```
-
-**A5.** We will use `{purrr}` package to apply this function. Key thing to keep in mind is that a data frame is a list of atomic vectors of equal length.
-
-- Applying function to every column in a data frame: We will use `anscombe` as example since it has all numeric columns.
-
-
-```r
-purrr::map_df(head(anscombe), .f = scale01)
-#> # A tibble: 6 × 8
-#> x1 x2 x3 x4 y1 y2 y3 y4
-#>
-
----
-
-**Q5.** The following code uses a map nested inside another map to apply a function to every element of a nested list. Why does it fail, and what do you need to do to make it work?
-
-
-```r
-x <- list(
- list(1, c(3, 9)),
- list(c(3, 6), 7, c(4, 7, 6))
-)
-
-triple <- function(x) x * 3
-map(x, map, .f = triple)
-#> Error in .f(.x[[i]], ...): unused argument (function (.x, .f, ...)
-#> {
-#> .f <- as_mapper(.f, ...)
-#> .Call(map_impl, environment(), ".x", ".f", "list")
-#> })
-```
-
-**A5.** This function fails because this call effectively evaluates to the following:
-
-
-```r
-map(.x = x, .f = ~ triple(x = .x, map))
-```
-
-But `triple()` has only one parameter (`x`), and so the execution fails.
-
-Here is the fixed version:
-
-
-```r
-x <- list(
- list(1, c(3, 9)),
- list(c(3, 6), 7, c(4, 7, 6))
-)
-
-triple <- function(x) x * 3
-map(x, .f = ~ map(.x, ~ triple(.x)))
-#> [[1]]
-#> [[1]][[1]]
-#> [1] 3
-#>
-#> [[1]][[2]]
-#> [1] 9 27
-#>
-#>
-#> [[2]]
-#> [[2]][[1]]
-#> [1] 9 18
-#>
-#> [[2]][[2]]
-#> [1] 21
-#>
-#> [[2]][[3]]
-#> [1] 12 21 18
-```
-
----
-
-**Q6.** Use `map()` to fit linear models to the `mtcars` dataset using the formulas stored in this list:
-
-
-```r
-formulas <- list(
- mpg ~ disp,
- mpg ~ I(1 / disp),
- mpg ~ disp + wt,
- mpg ~ I(1 / disp) + wt
-)
-```
-
-**A6.** Fitting linear models to the `mtcars` dataset using the provided formulas:
-
-
-```r
-formulas <- list(
- mpg ~ disp,
- mpg ~ I(1 / disp),
- mpg ~ disp + wt,
- mpg ~ I(1 / disp) + wt
-)
-
-map(formulas, ~ lm(formula = ., data = mtcars))
-#> [[1]]
-#>
-#> Call:
-#> lm(formula = ., data = mtcars)
-#>
-#> Coefficients:
-#> (Intercept) disp
-#> 29.59985 -0.04122
-#>
-#>
-#> [[2]]
-#>
-#> Call:
-#> lm(formula = ., data = mtcars)
-#>
-#> Coefficients:
-#> (Intercept) I(1/disp)
-#> 10.75 1557.67
-#>
-#>
-#> [[3]]
-#>
-#> Call:
-#> lm(formula = ., data = mtcars)
-#>
-#> Coefficients:
-#> (Intercept) disp wt
-#> 34.96055 -0.01772 -3.35083
-#>
-#>
-#> [[4]]
-#>
-#> Call:
-#> lm(formula = ., data = mtcars)
-#>
-#> Coefficients:
-#> (Intercept) I(1/disp) wt
-#> 19.024 1142.560 -1.798
-```
-
----
-
-**Q7.** Fit the model `mpg ~ disp` to each of the bootstrap replicates of `mtcars` in the list below, then extract the $R^2$ of the model fit (Hint: you can compute the $R^2$ with `summary()`.)
-
-
-```r
-bootstrap <- function(df) {
- df[sample(nrow(df), replace = TRUE), , drop = FALSE]
-}
-
-bootstraps <- map(1:10, ~ bootstrap(mtcars))
-```
-
-**A7.** This can be done using `map_dbl()`:
-
-
-```r
-bootstrap <- function(df) {
- df[sample(nrow(df), replace = TRUE), , drop = FALSE]
-}
-
-bootstraps <- map(1:10, ~ bootstrap(mtcars))
-
-bootstraps %>%
- map(~ lm(mpg ~ disp, data = .x)) %>%
- map(summary) %>%
- map_dbl("r.squared")
-#> [1] 0.7864562 0.8110818 0.7956331 0.7632399 0.7967824
-#> [6] 0.7364226 0.7203027 0.6653252 0.7732780 0.6753329
-```
-
----
-
-## Map variants (Exercises 9.4.6)
-
----
-
-**Q1.** Explain the results of `modify(mtcars, 1)`.
-
-**A1.** `modify()` returns the object of type same as the input. Since the input here is a data frame of certain dimensions and `.f = 1` translates to plucking the first element in each column, it returns a data frame with the same dimensions with the plucked element recycled across rows.
-
-
-```r
-head(modify(mtcars, 1))
-#> mpg cyl disp hp drat wt qsec vs am
-#> Mazda RX4 21 6 160 110 3.9 2.62 16.46 0 1
-#> Mazda RX4 Wag 21 6 160 110 3.9 2.62 16.46 0 1
-#> Datsun 710 21 6 160 110 3.9 2.62 16.46 0 1
-#> Hornet 4 Drive 21 6 160 110 3.9 2.62 16.46 0 1
-#> Hornet Sportabout 21 6 160 110 3.9 2.62 16.46 0 1
-#> Valiant 21 6 160 110 3.9 2.62 16.46 0 1
-#> gear carb
-#> Mazda RX4 4 4
-#> Mazda RX4 Wag 4 4
-#> Datsun 710 4 4
-#> Hornet 4 Drive 4 4
-#> Hornet Sportabout 4 4
-#> Valiant 4 4
-```
-
----
-
-**Q2.** Rewrite the following code to use `iwalk()` instead of `walk2()`. What are the advantages and disadvantages?
-
-
-```r
-cyls <- split(mtcars, mtcars$cyl)
-paths <- file.path(temp, paste0("cyl-", names(cyls), ".csv"))
-walk2(cyls, paths, write.csv)
-```
-
-**A2.** Let's first rewrite provided code using `iwalk()`:
-
-
-```r
-cyls <- split(mtcars, mtcars$cyl)
-names(cyls) <- file.path(temp, paste0("cyl-", names(cyls), ".csv"))
-iwalk(cyls, ~ write.csv(.x, .y))
-```
-
-The advantage of using `iwalk()` is that we need to now deal with only a single variable (`cyls`) instead of two (`cyls` and `paths`).
-
-The disadvantage is that the code is difficult to reason about:
-In `walk2()`, it's explicit what `.x` (`= cyls`) and `.y` (`= paths`) correspond to, while this is not so for `iwalk()` (i.e., `.x = cyls` and `.y = names(cyls)`) with the `.y` argument being "invisible".
-
----
-
-**Q3.** Explain how the following code transforms a data frame using functions stored in a list.
-
-
-```r
-trans <- list(
- disp = function(x) x * 0.0163871,
- am = function(x) factor(x, labels = c("auto", "manual"))
-)
-
-nm <- names(trans)
-mtcars[nm] <- map2(trans, mtcars[nm], function(f, var) f(var))
-```
-
-Compare and contrast the `map2()` approach to this `map()` approach:
-
-
-```r
-mtcars[nm] <- map(nm, ~ trans[[.x]](mtcars[[.x]]))
-```
-
-**A3.** `map2()` supplies the functions stored in `trans` as anonymous functions via placeholder `f`, while the names of the columns specified in `mtcars[nm]` are supplied as `var` argument to the anonymous function. Note that the function is iterating over indices for vectors of transformations and column names.
-
-
-```r
-trans <- list(
- disp = function(x) x * 0.0163871,
- am = function(x) factor(x, labels = c("auto", "manual"))
-)
-
-nm <- names(trans)
-mtcars[nm] <- map2(trans, mtcars[nm], function(f, var) f(var))
-```
-
-In the `map()` approach, the function is iterating over indices for vectors of column names.
-
-
-```r
-mtcars[nm] <- map(nm, ~ trans[[.x]](mtcars[[.x]]))
-```
-
-The latter approach can't afford passing arguments to placeholders in an anonymous function.
-
----
-
-**Q4.** What does `write.csv()` return, i.e. what happens if you use it with `map2()` instead of `walk2()`?
-
-**A4.** If we use `map2()`, it will work, but it will print `NULL`s to the console for every list element.
-
-
-```r
-withr::with_tempdir(
- code = {
- ls <- split(mtcars, mtcars$cyl)
- nm <- names(ls)
- map2(ls, nm, write.csv)
- }
-)
-#> $`4`
-#> NULL
-#>
-#> $`6`
-#> NULL
-#>
-#> $`8`
-#> NULL
-```
-
----
-
-## Predicate functionals (Exercises 9.6.3)
-
----
-
-**Q1.** Why isn't `is.na()` a predicate function? What base R function is closest to being a predicate version of `is.na()`?
-
-**A1.** As mentioned in the docs:
-
-> A predicate is a function that returns a **single** `TRUE` or `FALSE`.
-
-The `is.na()` function does not return a `logical` scalar, but instead returns a vector and thus isn't a predicate function.
-
-
-```r
-# contrast the following behavior of predicate functions
-is.character(c("x", 2))
-#> [1] TRUE
-is.null(c(3, NULL))
-#> [1] FALSE
-
-# with this behavior
-is.na(c(NA, 1))
-#> [1] TRUE FALSE
-```
-
-The closest equivalent of a predicate function in base-R is `anyNA()` function.
-
-
-```r
-anyNA(c(NA, 1))
-#> [1] TRUE
-```
-
----
-
-**Q2.** `simple_reduce()` has a problem when `x` is length 0 or length 1. Describe the source of the problem and how you might go about fixing it.
-
-
-```r
-simple_reduce <- function(x, f) {
- out <- x[[1]]
- for (i in seq(2, length(x))) {
- out <- f(out, x[[i]])
- }
- out
-}
-```
-
-**A2.** The supplied function struggles with inputs of length 0 and 1 because function tries to subscript out-of-bound values.
-
-
-```r
-simple_reduce(numeric(), sum)
-#> Error in x[[1]]: subscript out of bounds
-simple_reduce(1, sum)
-#> Error in x[[i]]: subscript out of bounds
-simple_reduce(1:3, sum)
-#> [1] 6
-```
-
-This problem can be solved by adding `init` argument, which supplies the default or initial value:
-
-
-```r
-simple_reduce2 <- function(x, f, init = 0) {
- # initializer will become the first value
- if (length(x) == 0L) {
- return(init)
- }
-
- if (length(x) == 1L) {
- return(x[[1L]])
- }
-
- out <- x[[1]]
-
- for (i in seq(2, length(x))) {
- out <- f(out, x[[i]])
- }
-
- out
-}
-```
-
-Let's try it out:
-
-
-```r
-simple_reduce2(numeric(), sum)
-#> [1] 0
-simple_reduce2(1, sum)
-#> [1] 1
-simple_reduce2(1:3, sum)
-#> [1] 6
-```
-
-Depending on the function, we can provide a different `init` argument:
-
-
-```r
-simple_reduce2(numeric(), `*`, init = 1)
-#> [1] 1
-simple_reduce2(1, `*`, init = 1)
-#> [1] 1
-simple_reduce2(1:3, `*`, init = 1)
-#> [1] 6
-```
-
----
-
-**Q3.** Implement the `span()` function from Haskell: given a list `x` and a predicate function `f`, `span(x, f)` returns the location of the longest sequential run of elements where the predicate is true. (Hint: you might find `rle()` helpful.)
-
-**A3.** Implementation of `span()`:
-
-
-```r
-span <- function(x, f) {
- running_lengths <- purrr::map_lgl(x, ~ f(.x)) %>% rle()
-
- df <- dplyr::tibble(
- "lengths" = running_lengths$lengths,
- "values" = running_lengths$values
- ) %>%
- dplyr::mutate(rowid = dplyr::row_number()) %>%
- dplyr::filter(values)
-
- # no sequence where condition is `TRUE`
- if (nrow(df) == 0L) {
- return(integer())
- }
-
- # only single sequence where condition is `TRUE`
- if (nrow(df) == 1L) {
- return((df$rowid):(df$lengths - 1 + df$rowid))
- }
-
- # multiple sequences where condition is `TRUE`; select max one
- if (nrow(df) > 1L) {
- df <- dplyr::filter(df, lengths == max(lengths))
- return((df$rowid):(df$lengths - 1 + df$rowid))
- }
-}
-```
-
-Testing it once:
-
-
-```r
-span(c(0, 0, 0, 0, 0), is.na)
-#> integer(0)
-span(c(NA, 0, NA, NA, NA), is.na)
-#> [1] 3 4 5
-span(c(NA, 0, 0, 0, 0), is.na)
-#> [1] 1
-span(c(NA, NA, 0, 0, 0), is.na)
-#> [1] 1 2
-```
-
-Testing it twice:
-
-
-```r
-span(c(3, 1, 2, 4, 5, 6), function(x) x > 3)
-#> [1] 2 3 4
-span(c(3, 1, 2, 4, 5, 6), function(x) x > 9)
-#> integer(0)
-span(c(3, 1, 2, 4, 5, 6), function(x) x == 3)
-#> [1] 1
-span(c(3, 1, 2, 4, 5, 6), function(x) x %in% c(2, 4))
-#> [1] 2 3
-```
-
----
-
-**Q4.** Implement `arg_max()`. It should take a function and a vector of inputs, and return the elements of the input where the function returns the highest value. For example, `arg_max(-10:5, function(x) x ^ 2)` should return -10. `arg_max(-5:5, function(x) x ^ 2)` should return `c(-5, 5)`. Also implement the matching `arg_min()` function.
-
-**A4.** Here are implementations for the specified functions:
-
-- Implementing `arg_max()`
-
-
-```r
-arg_max <- function(.x, .f) {
- df <- dplyr::tibble(
- original = .x,
- transformed = purrr::map_dbl(.x, .f)
- )
-
- dplyr::filter(df, transformed == max(transformed))[["original"]]
-}
-
-arg_max(-10:5, function(x) x^2)
-#> [1] -10
-arg_max(-5:5, function(x) x^2)
-#> [1] -5 5
-```
-
-- Implementing `arg_min()`
-
-
-```r
-arg_min <- function(.x, .f) {
- df <- dplyr::tibble(
- original = .x,
- transformed = purrr::map_dbl(.x, .f)
- )
-
- dplyr::filter(df, transformed == min(transformed))[["original"]]
-}
-
-arg_min(-10:5, function(x) x^2)
-#> [1] 0
-arg_min(-5:5, function(x) x^2)
-#> [1] 0
-```
-
----
-
-**Q5.** The function below scales a vector so it falls in the range [0, 1]. How would you apply it to every column of a data frame? How would you apply it to every numeric column in a data frame?
-
-
-```r
-scale01 <- function(x) {
- rng <- range(x, na.rm = TRUE)
- (x - rng[1]) / (rng[2] - rng[1])
-}
-```
-
-**A5.** We will use `{purrr}` package to apply this function. Key thing to keep in mind is that a data frame is a list of atomic vectors of equal length.
-
-- Applying function to every column in a data frame: We will use `anscombe` as example since it has all numeric columns.
-
-
-```r
-purrr::map_df(head(anscombe), .f = scale01)
-#> # A tibble: 6 × 8
-#> x1 x2 x3 x4 y1 y2 y3 y4
-#>  -
-The default `xlim = range(breaks)` and `breaks = "Sturges"` arguments reveal that the function uses Sturges' algorithm to compute the number of breaks.
-
-
-```r
-nclass.Sturges(mtcars$wt)
-#> [1] 6
-```
-
-To see the implementation, run `sloop::s3_get_method("hist.default")`.
-
-`hist()` ensures that the chosen algorithm returns a numeric vector containing at least two unique elements before `xlim` is computed.
-
-**Q5.** Explain why this function works. Why is it confusing?
-
-
-```r
-show_time <- function(x = stop("Error!")) {
- stop <- function(...) Sys.time()
- print(x)
-}
-
-show_time()
-#> [1] "2022-11-12 11:48:38 CET"
-```
-
-**A5.** Let's take this step-by-step.
-
-The function argument `x` is missing in the function call. This means that `stop("Error!")` is evaluated in the function environment, and not global environment.
-
-But, due to lazy evaluation, the promise `stop("Error!")` is evaluated only when `x` is accessed. This happens only when `print(x)` is called.
-
-`print(x)` leads to `x` being evaluated, which evaluates `stop` in the function environment. But, in function environment, the `base::stop()` is masked by a locally defined `stop()` function, which returns `Sys.time()` output.
-
-**Q6.** How many arguments are required when calling `library()`?
-
-**A6.** Going solely by its signature,
-
-
-```r
-formals(library)
-#> $package
-#>
-#>
-#> $help
-#>
-#>
-#> $pos
-#> [1] 2
-#>
-#> $lib.loc
-#> NULL
-#>
-#> $character.only
-#> [1] FALSE
-#>
-#> $logical.return
-#> [1] FALSE
-#>
-#> $warn.conflicts
-#>
-#>
-#> $quietly
-#> [1] FALSE
-#>
-#> $verbose
-#> getOption("verbose")
-#>
-#> $mask.ok
-#>
-#>
-#> $exclude
-#>
-#>
-#> $include.only
-#>
-#>
-#> $attach.required
-#> missing(include.only)
-```
-
-it looks like the following arguments are required:
-
-
-```r
-formals(library) %>%
- purrr::discard(is.null) %>%
- purrr::map_lgl(~ .x == "") %>%
- purrr::keep(~ isTRUE(.x)) %>%
- names()
-#> [1] "package" "help" "warn.conflicts"
-#> [4] "mask.ok" "exclude" "include.only"
-```
-
-But, in reality, only one argument is required: `package`. The function internally checks if the other arguments are missing and adjusts accordingly.
-
-It would have been better if there arguments were `NULL` instead of missing; that would avoid this confusion.
-
-## `...` (dot-dot-dot) (Exercises 6.6.1)
-
-**Q1.** Explain the following results:
-
-
-```r
-sum(1, 2, 3)
-#> [1] 6
-mean(1, 2, 3)
-#> [1] 1
-
-sum(1, 2, 3, na.omit = TRUE)
-#> [1] 7
-mean(1, 2, 3, na.omit = TRUE)
-#> [1] 1
-```
-
-**A1.** Let's look at arguments for these functions:
-
-
-```r
-str(sum)
-#> function (..., na.rm = FALSE)
-str(mean)
-#> function (x, ...)
-```
-
-As can be seen, `sum()` function doesn't have `na.omit` argument. So, the input `na.omit = TRUE` is treated as `1` (logical implicitly coerced to numeric), and thus the results. So, the expression evaluates to `sum(1, 2, 3, 1)`.
-
-For `mean()` function, there is only one parameter (`x`) and it's matched by the first argument (`1`). So, the expression evaluates to `mean(1)`.
-
-**Q2.** Explain how to find the documentation for the named arguments in the following function call:
-
-
-```r
-plot(1:10, col = "red", pch = 20, xlab = "x", col.lab = "blue")
-```
-
-
-
-The default `xlim = range(breaks)` and `breaks = "Sturges"` arguments reveal that the function uses Sturges' algorithm to compute the number of breaks.
-
-
-```r
-nclass.Sturges(mtcars$wt)
-#> [1] 6
-```
-
-To see the implementation, run `sloop::s3_get_method("hist.default")`.
-
-`hist()` ensures that the chosen algorithm returns a numeric vector containing at least two unique elements before `xlim` is computed.
-
-**Q5.** Explain why this function works. Why is it confusing?
-
-
-```r
-show_time <- function(x = stop("Error!")) {
- stop <- function(...) Sys.time()
- print(x)
-}
-
-show_time()
-#> [1] "2022-11-12 11:48:38 CET"
-```
-
-**A5.** Let's take this step-by-step.
-
-The function argument `x` is missing in the function call. This means that `stop("Error!")` is evaluated in the function environment, and not global environment.
-
-But, due to lazy evaluation, the promise `stop("Error!")` is evaluated only when `x` is accessed. This happens only when `print(x)` is called.
-
-`print(x)` leads to `x` being evaluated, which evaluates `stop` in the function environment. But, in function environment, the `base::stop()` is masked by a locally defined `stop()` function, which returns `Sys.time()` output.
-
-**Q6.** How many arguments are required when calling `library()`?
-
-**A6.** Going solely by its signature,
-
-
-```r
-formals(library)
-#> $package
-#>
-#>
-#> $help
-#>
-#>
-#> $pos
-#> [1] 2
-#>
-#> $lib.loc
-#> NULL
-#>
-#> $character.only
-#> [1] FALSE
-#>
-#> $logical.return
-#> [1] FALSE
-#>
-#> $warn.conflicts
-#>
-#>
-#> $quietly
-#> [1] FALSE
-#>
-#> $verbose
-#> getOption("verbose")
-#>
-#> $mask.ok
-#>
-#>
-#> $exclude
-#>
-#>
-#> $include.only
-#>
-#>
-#> $attach.required
-#> missing(include.only)
-```
-
-it looks like the following arguments are required:
-
-
-```r
-formals(library) %>%
- purrr::discard(is.null) %>%
- purrr::map_lgl(~ .x == "") %>%
- purrr::keep(~ isTRUE(.x)) %>%
- names()
-#> [1] "package" "help" "warn.conflicts"
-#> [4] "mask.ok" "exclude" "include.only"
-```
-
-But, in reality, only one argument is required: `package`. The function internally checks if the other arguments are missing and adjusts accordingly.
-
-It would have been better if there arguments were `NULL` instead of missing; that would avoid this confusion.
-
-## `...` (dot-dot-dot) (Exercises 6.6.1)
-
-**Q1.** Explain the following results:
-
-
-```r
-sum(1, 2, 3)
-#> [1] 6
-mean(1, 2, 3)
-#> [1] 1
-
-sum(1, 2, 3, na.omit = TRUE)
-#> [1] 7
-mean(1, 2, 3, na.omit = TRUE)
-#> [1] 1
-```
-
-**A1.** Let's look at arguments for these functions:
-
-
-```r
-str(sum)
-#> function (..., na.rm = FALSE)
-str(mean)
-#> function (x, ...)
-```
-
-As can be seen, `sum()` function doesn't have `na.omit` argument. So, the input `na.omit = TRUE` is treated as `1` (logical implicitly coerced to numeric), and thus the results. So, the expression evaluates to `sum(1, 2, 3, 1)`.
-
-For `mean()` function, there is only one parameter (`x`) and it's matched by the first argument (`1`). So, the expression evaluates to `mean(1)`.
-
-**Q2.** Explain how to find the documentation for the named arguments in the following function call:
-
-
-```r
-plot(1:10, col = "red", pch = 20, xlab = "x", col.lab = "blue")
-```
-
- -
-**A2.** Typing `?plot` in the console, we see its documentation, which also shows its signature:
-
-
-```
-#> function (x, y, ...)
-```
-
-Since `...` are passed to `par()`, we can look at `?par` docs:
-
-
-```
-#> function (..., no.readonly = FALSE)
-```
-
-And so on.
-
-The docs for all parameters of interest [reside there](https://rdrr.io/r/graphics/par.html).
-
-**Q3.** Why does `plot(1:10, col = "red")` only colour the points, not the axes or labels? Read the source code of `plot.default()` to find out.
-
-**A3.** Source code can be found [here](https://github.com/wch/r-source/blob/79e73dba5259b25ec30118d45fea64aeac0f41dc/src/library/graphics/R/plot.R#L51-L84).
-
-`plot.default()` passes `...` to `localTitle()`, which passes it to `title()`.
-
-`title()` has four parts: `main`, `sub`, `xlab`, `ylab`.
-
-So having a single argument `col` would not work as it will be ambiguous as to which element to apply this argument to.
-
-
-```r
-localTitle <- function(..., col, bg, pch, cex, lty, lwd) title(...)
-
-title <- function(main = NULL, sub = NULL, xlab = NULL, ylab = NULL,
- line = NA, outer = FALSE, ...) {
- main <- as.graphicsAnnot(main)
- sub <- as.graphicsAnnot(sub)
- xlab <- as.graphicsAnnot(xlab)
- ylab <- as.graphicsAnnot(ylab)
- .External.graphics(C_title, main, sub, xlab, ylab, line, outer, ...)
- invisible()
-}
-```
-
-## Exiting a function (Exercises 6.7.5)
-
-**Q1.** What does `load()` return? Why don't you normally see these values?
-
-**A1.** The `load()` function reloads datasets that were saved using the `save()` function:
-
-
-```r
-save(iris, file = "my_iris.rda")
-load("my_iris.rda")
-```
-
-We normally don't see any value because the function loads the datasets invisibly.
-
-We can change this by setting `verbose = TRUE`:
-
-
-```r
-load("my_iris.rda", verbose = TRUE)
-#> Loading objects:
-#> iris
-
-# cleanup
-unlink("my_iris.rda")
-```
-
-**Q2.** What does `write.table()` return? What would be more useful?
-
-**A2.** The `write.table()` writes a data frame to a file and returns a `NULL` invisibly.
-
-
-```r
-write.table(BOD, file = "BOD.csv")
-```
-
-It would have been more helpful if the function invisibly returned the actual object being written to the file, which could then be further used.
-
-
-```r
-# cleanup
-unlink("BOD.csv")
-```
-
-**Q3.** How does the `chdir` parameter of `source()` compare to `with_dir()`? Why might you prefer one to the other?
-
-**A3.** The `chdir` parameter of `source()` is described as:
-
-> if `TRUE` and `file` is a pathname, the `R` working directory is temporarily changed to the directory containing file for evaluating
-
-That is, `chdir` allows changing working directory temporarily but *only* to the directory containing file being sourced:
-
-While `withr::with_dir()` temporarily changes the current working directory:
-
-
-```r
-withr::with_dir
-#> function (new, code)
-#> {
-#> old <- setwd(dir = new)
-#> on.exit(setwd(old))
-#> force(code)
-#> }
-#>
-
-**A2.** Typing `?plot` in the console, we see its documentation, which also shows its signature:
-
-
-```
-#> function (x, y, ...)
-```
-
-Since `...` are passed to `par()`, we can look at `?par` docs:
-
-
-```
-#> function (..., no.readonly = FALSE)
-```
-
-And so on.
-
-The docs for all parameters of interest [reside there](https://rdrr.io/r/graphics/par.html).
-
-**Q3.** Why does `plot(1:10, col = "red")` only colour the points, not the axes or labels? Read the source code of `plot.default()` to find out.
-
-**A3.** Source code can be found [here](https://github.com/wch/r-source/blob/79e73dba5259b25ec30118d45fea64aeac0f41dc/src/library/graphics/R/plot.R#L51-L84).
-
-`plot.default()` passes `...` to `localTitle()`, which passes it to `title()`.
-
-`title()` has four parts: `main`, `sub`, `xlab`, `ylab`.
-
-So having a single argument `col` would not work as it will be ambiguous as to which element to apply this argument to.
-
-
-```r
-localTitle <- function(..., col, bg, pch, cex, lty, lwd) title(...)
-
-title <- function(main = NULL, sub = NULL, xlab = NULL, ylab = NULL,
- line = NA, outer = FALSE, ...) {
- main <- as.graphicsAnnot(main)
- sub <- as.graphicsAnnot(sub)
- xlab <- as.graphicsAnnot(xlab)
- ylab <- as.graphicsAnnot(ylab)
- .External.graphics(C_title, main, sub, xlab, ylab, line, outer, ...)
- invisible()
-}
-```
-
-## Exiting a function (Exercises 6.7.5)
-
-**Q1.** What does `load()` return? Why don't you normally see these values?
-
-**A1.** The `load()` function reloads datasets that were saved using the `save()` function:
-
-
-```r
-save(iris, file = "my_iris.rda")
-load("my_iris.rda")
-```
-
-We normally don't see any value because the function loads the datasets invisibly.
-
-We can change this by setting `verbose = TRUE`:
-
-
-```r
-load("my_iris.rda", verbose = TRUE)
-#> Loading objects:
-#> iris
-
-# cleanup
-unlink("my_iris.rda")
-```
-
-**Q2.** What does `write.table()` return? What would be more useful?
-
-**A2.** The `write.table()` writes a data frame to a file and returns a `NULL` invisibly.
-
-
-```r
-write.table(BOD, file = "BOD.csv")
-```
-
-It would have been more helpful if the function invisibly returned the actual object being written to the file, which could then be further used.
-
-
-```r
-# cleanup
-unlink("BOD.csv")
-```
-
-**Q3.** How does the `chdir` parameter of `source()` compare to `with_dir()`? Why might you prefer one to the other?
-
-**A3.** The `chdir` parameter of `source()` is described as:
-
-> if `TRUE` and `file` is a pathname, the `R` working directory is temporarily changed to the directory containing file for evaluating
-
-That is, `chdir` allows changing working directory temporarily but *only* to the directory containing file being sourced:
-
-While `withr::with_dir()` temporarily changes the current working directory:
-
-
-```r
-withr::with_dir
-#> function (new, code)
-#> {
-#> old <- setwd(dir = new)
-#> on.exit(setwd(old))
-#> force(code)
-#> }
-#>  -
----
-
-## Object size (Exercise 2.4.1)
-
----
-
-**Q1.** In the following example, why are `object.size(y)` and `obj_size(y)` so radically different? Consult the documentation of `object.size()`.
-
-
-```r
-y <- rep(list(runif(1e4)), 100)
-
-object.size(y)
-obj_size(y)
-```
-
-**A1.** As mentioned in the docs for `object.size()`:
-
-> This function...does not detect if elements of a list are shared.
-
-This is why the sizes are so different:
-
-
-```r
-y <- rep(list(runif(1e4)), 100)
-
-object.size(y)
-#> 8005648 bytes
-
-obj_size(y)
-#> 80.90 kB
-```
-
----
-
-**Q2.** Take the following list. Why is its size somewhat misleading?
-
-
-```r
-funs <- list(mean, sd, var)
-obj_size(funs)
-```
-
-**A2.** These functions are not externally created objects in R, but are always available as part of base packages, so doesn't make much sense to measure their size because they are never going to be *not* available.
-
-
-```r
-funs <- list(mean, sd, var)
-obj_size(funs)
-#> 17.55 kB
-```
-
----
-
-**Q3.** Predict the output of the following code:
-
-
-```r
-a <- runif(1e6)
-obj_size(a)
-
-b <- list(a, a)
-obj_size(b)
-obj_size(a, b)
-
-b[[1]][[1]] <- 10
-obj_size(b)
-obj_size(a, b)
-
-b[[2]][[1]] <- 10
-obj_size(b)
-obj_size(a, b)
-```
-
-**A3.** Correctly predicted 😉
-
-
-```r
-a <- runif(1e6)
-obj_size(a)
-#> 8.00 MB
-
-b <- list(a, a)
-obj_size(b)
-#> 8.00 MB
-obj_size(a, b)
-#> 8.00 MB
-
-b[[1]][[1]] <- 10
-obj_size(b)
-#> 16.00 MB
-obj_size(a, b)
-#> 16.00 MB
-
-b[[2]][[1]] <- 10
-obj_size(b)
-#> 16.00 MB
-obj_size(a, b)
-#> 24.00 MB
-```
-
-Key pieces of information to keep in mind to make correct predictions:
-
-- Size of empty vector
-
-
-```r
-obj_size(double())
-#> 48 B
-```
-
-- Size of a single double: 8 bytes
-
-
-```r
-obj_size(double(1))
-#> 56 B
-```
-
-- Copy-on-modify semantics
-
----
-
-## Modify-in-place (Exercise 2.5.3)
-
----
-
-**Q1.** Explain why the following code doesn't create a circular list.
-
-
-```r
-x <- list()
-x[[1]] <- x
-```
-
-**A1.** Copy-on-modify prevents the creation of a circular list.
-
-
-```r
-x <- list()
-
-obj_addr(x)
-#> [1] "0x106c2ac38"
-
-tracemem(x)
-#> [1] "<0x106c2ac38>"
-
-x[[1]] <- x
-#> tracemem[0x106c2ac38 -> 0x12a7d3a50]: eval eval eval_with_user_handlers withVisible withCallingHandlers handle timing_fn evaluate_call
-
----
-
-## Object size (Exercise 2.4.1)
-
----
-
-**Q1.** In the following example, why are `object.size(y)` and `obj_size(y)` so radically different? Consult the documentation of `object.size()`.
-
-
-```r
-y <- rep(list(runif(1e4)), 100)
-
-object.size(y)
-obj_size(y)
-```
-
-**A1.** As mentioned in the docs for `object.size()`:
-
-> This function...does not detect if elements of a list are shared.
-
-This is why the sizes are so different:
-
-
-```r
-y <- rep(list(runif(1e4)), 100)
-
-object.size(y)
-#> 8005648 bytes
-
-obj_size(y)
-#> 80.90 kB
-```
-
----
-
-**Q2.** Take the following list. Why is its size somewhat misleading?
-
-
-```r
-funs <- list(mean, sd, var)
-obj_size(funs)
-```
-
-**A2.** These functions are not externally created objects in R, but are always available as part of base packages, so doesn't make much sense to measure their size because they are never going to be *not* available.
-
-
-```r
-funs <- list(mean, sd, var)
-obj_size(funs)
-#> 17.55 kB
-```
-
----
-
-**Q3.** Predict the output of the following code:
-
-
-```r
-a <- runif(1e6)
-obj_size(a)
-
-b <- list(a, a)
-obj_size(b)
-obj_size(a, b)
-
-b[[1]][[1]] <- 10
-obj_size(b)
-obj_size(a, b)
-
-b[[2]][[1]] <- 10
-obj_size(b)
-obj_size(a, b)
-```
-
-**A3.** Correctly predicted 😉
-
-
-```r
-a <- runif(1e6)
-obj_size(a)
-#> 8.00 MB
-
-b <- list(a, a)
-obj_size(b)
-#> 8.00 MB
-obj_size(a, b)
-#> 8.00 MB
-
-b[[1]][[1]] <- 10
-obj_size(b)
-#> 16.00 MB
-obj_size(a, b)
-#> 16.00 MB
-
-b[[2]][[1]] <- 10
-obj_size(b)
-#> 16.00 MB
-obj_size(a, b)
-#> 24.00 MB
-```
-
-Key pieces of information to keep in mind to make correct predictions:
-
-- Size of empty vector
-
-
-```r
-obj_size(double())
-#> 48 B
-```
-
-- Size of a single double: 8 bytes
-
-
-```r
-obj_size(double(1))
-#> 56 B
-```
-
-- Copy-on-modify semantics
-
----
-
-## Modify-in-place (Exercise 2.5.3)
-
----
-
-**Q1.** Explain why the following code doesn't create a circular list.
-
-
-```r
-x <- list()
-x[[1]] <- x
-```
-
-**A1.** Copy-on-modify prevents the creation of a circular list.
-
-
-```r
-x <- list()
-
-obj_addr(x)
-#> [1] "0x106c2ac38"
-
-tracemem(x)
-#> [1] "<0x106c2ac38>"
-
-x[[1]] <- x
-#> tracemem[0x106c2ac38 -> 0x12a7d3a50]: eval eval eval_with_user_handlers withVisible withCallingHandlers handle timing_fn evaluate_call  -
----
-
-**Q3.** What happens if you attempt to use `tracemem()` on an environment?
-
-**A3.** It doesn't work and the documentation for `tracemem()` makes it clear why:
-
-> It is not useful to trace `NULL`, environments, promises, weak references, or external pointer objects, as these are not duplicated
-
-
-```r
-e <- rlang::env(a = 1, b = "3")
-tracemem(e)
-#> Error in tracemem(e): 'tracemem' is not useful for promise and environment objects
-```
-
----
-
-## Session information
-
-
-```r
-sessioninfo::session_info(include_base = TRUE)
-#> ─ Session info ───────────────────────────────────────────
-#> setting value
-#> version R version 4.2.2 (2022-10-31)
-#> os macOS Ventura 13.0
-#> system aarch64, darwin20
-#> ui X11
-#> language (EN)
-#> collate en_US.UTF-8
-#> ctype en_US.UTF-8
-#> tz Europe/Berlin
-#> date 2022-11-12
-#> pandoc 2.19.2 @ /Applications/RStudio.app/Contents/MacOS/quarto/bin/tools/ (via rmarkdown)
-#>
-#> ─ Packages ───────────────────────────────────────────────
-#> ! package * version date (UTC) lib source
-#> assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.2.0)
-#> backports 1.4.1 2021-12-13 [1] CRAN (R 4.2.0)
-#> base * 4.2.2 2022-10-31 [?] local
-#> bench * 1.1.2 2021-11-30 [1] CRAN (R 4.2.0)
-#> bookdown 0.30 2022-11-09 [1] CRAN (R 4.2.2)
-#> broom 1.0.1 2022-08-29 [1] CRAN (R 4.2.0)
-#> bslib 0.4.1 2022-11-02 [1] CRAN (R 4.2.2)
-#> cachem 1.0.6 2021-08-19 [1] CRAN (R 4.2.0)
-#> cellranger 1.1.0 2016-07-27 [1] CRAN (R 4.2.0)
-#> cli 3.4.1 2022-09-23 [1] CRAN (R 4.2.0)
-#> colorspace 2.0-3 2022-02-21 [1] CRAN (R 4.2.0)
-#> P compiler 4.2.2 2022-10-31 [1] local
-#> crayon 1.5.2 2022-09-29 [1] CRAN (R 4.2.1)
-#> P datasets * 4.2.2 2022-10-31 [1] local
-#> DBI 1.1.3.9002 2022-10-17 [1] Github (r-dbi/DBI@2aec388)
-#> dbplyr 2.2.1 2022-06-27 [1] CRAN (R 4.2.0)
-#> digest 0.6.30 2022-10-18 [1] CRAN (R 4.2.1)
-#> downlit 0.4.2 2022-07-05 [1] CRAN (R 4.2.1)
-#> dplyr * 1.0.10 2022-09-01 [1] CRAN (R 4.2.1)
-#> ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.2.0)
-#> evaluate 0.18 2022-11-07 [1] CRAN (R 4.2.2)
-#> fansi 1.0.3 2022-03-24 [1] CRAN (R 4.2.0)
-#> farver 2.1.1 2022-07-06 [1] CRAN (R 4.2.1)
-#> fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.2.0)
-#> forcats * 0.5.2 2022-08-19 [1] CRAN (R 4.2.1)
-#> fs 1.5.2 2021-12-08 [1] CRAN (R 4.2.0)
-#> gargle 1.2.1 2022-09-08 [1] CRAN (R 4.2.1)
-#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.2.1)
-#> ggplot2 * 3.4.0 2022-11-04 [1] CRAN (R 4.2.2)
-#> glue 1.6.2 2022-02-24 [1] CRAN (R 4.2.0)
-#> googledrive 2.0.0 2021-07-08 [1] CRAN (R 4.2.0)
-#> googlesheets4 1.0.1 2022-08-13 [1] CRAN (R 4.2.0)
-#> P graphics * 4.2.2 2022-10-31 [1] local
-#> P grDevices * 4.2.2 2022-10-31 [1] local
-#> P grid 4.2.2 2022-10-31 [1] local
-#> gtable 0.3.1 2022-09-01 [1] CRAN (R 4.2.1)
-#> haven 2.5.1 2022-08-22 [1] CRAN (R 4.2.0)
-#> highr 0.9 2021-04-16 [1] CRAN (R 4.2.0)
-#> hms 1.1.2 2022-08-19 [1] CRAN (R 4.2.0)
-#> htmltools 0.5.3 2022-07-18 [1] CRAN (R 4.2.1)
-#> httr 1.4.4 2022-08-17 [1] CRAN (R 4.2.0)
-#> jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.2.0)
-#> jsonlite 1.8.3 2022-10-21 [1] CRAN (R 4.2.1)
-#> knitr 1.40 2022-08-24 [1] CRAN (R 4.2.1)
-#> labeling 0.4.2 2020-10-20 [1] CRAN (R 4.2.0)
-#> lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.2.1)
-#> lobstr * 1.1.2 2022-06-22 [1] CRAN (R 4.2.0)
-#> lubridate 1.9.0 2022-11-06 [1] CRAN (R 4.2.2)
-#> magrittr * 2.0.3 2022-03-30 [1] CRAN (R 4.2.0)
-#> memoise 2.0.1 2021-11-26 [1] CRAN (R 4.2.0)
-#> P methods * 4.2.2 2022-10-31 [1] local
-#> modelr 0.1.10 2022-11-11 [1] CRAN (R 4.2.2)
-#> munsell 0.5.0 2018-06-12 [1] CRAN (R 4.2.0)
-#> pillar 1.8.1 2022-08-19 [1] CRAN (R 4.2.1)
-#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.2.0)
-#> prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.2.0)
-#> profmem 0.6.0 2020-12-13 [1] CRAN (R 4.2.0)
-#> purrr * 0.3.5 2022-10-06 [1] CRAN (R 4.2.1)
-#> R6 2.5.1.9000 2022-10-27 [1] local
-#> readr * 2.1.3 2022-10-01 [1] CRAN (R 4.2.1)
-#> readxl 1.4.1 2022-08-17 [1] CRAN (R 4.2.0)
-#> reprex 2.0.2 2022-08-17 [1] CRAN (R 4.2.1)
-#> rlang 1.0.6 2022-09-24 [1] CRAN (R 4.2.1)
-#> rmarkdown 2.18 2022-11-09 [1] CRAN (R 4.2.2)
-#> rstudioapi 0.14 2022-08-22 [1] CRAN (R 4.2.1)
-#> rvest 1.0.3 2022-08-19 [1] CRAN (R 4.2.1)
-#> sass 0.4.2 2022-07-16 [1] CRAN (R 4.2.1)
-#> scales 1.2.1 2022-08-20 [1] CRAN (R 4.2.1)
-#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.2.0)
-#> P stats * 4.2.2 2022-10-31 [1] local
-#> stringi 1.7.8 2022-07-11 [1] CRAN (R 4.2.1)
-#> stringr * 1.4.1 2022-08-20 [1] CRAN (R 4.2.1)
-#> tibble * 3.1.8.9002 2022-10-16 [1] local
-#> tidyr * 1.2.1 2022-09-08 [1] CRAN (R 4.2.1)
-#> tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.2.1)
-#> tidyverse * 1.3.2 2022-07-18 [1] CRAN (R 4.2.0)
-#> timechange 0.1.1 2022-11-04 [1] CRAN (R 4.2.2)
-#> P tools 4.2.2 2022-10-31 [1] local
-#> tzdb 0.3.0 2022-03-28 [1] CRAN (R 4.2.0)
-#> utf8 1.2.2 2021-07-24 [1] CRAN (R 4.2.0)
-#> P utils * 4.2.2 2022-10-31 [1] local
-#> vctrs 0.5.0 2022-10-22 [1] CRAN (R 4.2.1)
-#> withr 2.5.0 2022-03-03 [1] CRAN (R 4.2.0)
-#> xfun 0.34 2022-10-18 [1] CRAN (R 4.2.1)
-#> xml2 1.3.3.9000 2022-10-10 [1] local
-#> yaml 2.3.6 2022-10-18 [1] CRAN (R 4.2.1)
-#>
-#> [1] /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library
-#>
-#> P ── Loaded and on-disk path mismatch.
-#>
-#> ──────────────────────────────────────────────────────────
-```
diff --git a/_book/Names-values_files/figure-html/Names-values-31-1.png b/_book/Names-values_files/figure-html/Names-values-31-1.png
deleted file mode 100644
index 6210e6f4..00000000
Binary files a/_book/Names-values_files/figure-html/Names-values-31-1.png and /dev/null differ
diff --git a/_book/OO-tradeoffs.md b/_book/OO-tradeoffs.md
deleted file mode 100644
index afc23c3c..00000000
--- a/_book/OO-tradeoffs.md
+++ /dev/null
@@ -1,3 +0,0 @@
-# Trade-offs
-
-No exercises.
diff --git a/_book/Perf-improve.md b/_book/Perf-improve.md
deleted file mode 100644
index c9dc4ff3..00000000
--- a/_book/Perf-improve.md
+++ /dev/null
@@ -1,606 +0,0 @@
-# Improving performance
-
-
-
-Attaching the needed libraries:
-
-
-```r
-library(ggplot2)
-library(dplyr)
-library(purrr)
-```
-
-## Exercises 24.3.1
-
-**Q1.** What are faster alternatives to `lm()`? Which are specifically designed to work with larger datasets?
-
-**A1.** Faster alternatives to `lm()` can be found by visiting [CRAN Task View: High-Performance and Parallel Computing with R](https://cran.r-project.org/web/views/HighPerformanceComputing.html) page.
-
-Here are some of the available options:
-
-- `speedglm::speedlm()` (for large datasets)
-
-- `biglm::biglm()` (specifically designed for data too large to fit in memory)
-
-- `RcppEigen::fastLm()` (using the `Eigen` linear algebra library)
-
-High performances can be obtained with these packages especially if R is linked against an optimized BLAS, such as ATLAS. You can check this information using `sessionInfo()`:
-
-
-```r
-sessInfo <- sessionInfo()
-sessInfo$matprod
-#> [1] "default"
-sessInfo$LAPACK
-#> [1] "/Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/lib/libRlapack.dylib"
-```
-
-Comparing performance of different alternatives:
-
-
-```r
-library(gapminder)
-
-# having a look at the data
-glimpse(gapminder)
-#> Rows: 1,704
-#> Columns: 6
-#> $ country
-
----
-
-**Q3.** What happens if you attempt to use `tracemem()` on an environment?
-
-**A3.** It doesn't work and the documentation for `tracemem()` makes it clear why:
-
-> It is not useful to trace `NULL`, environments, promises, weak references, or external pointer objects, as these are not duplicated
-
-
-```r
-e <- rlang::env(a = 1, b = "3")
-tracemem(e)
-#> Error in tracemem(e): 'tracemem' is not useful for promise and environment objects
-```
-
----
-
-## Session information
-
-
-```r
-sessioninfo::session_info(include_base = TRUE)
-#> ─ Session info ───────────────────────────────────────────
-#> setting value
-#> version R version 4.2.2 (2022-10-31)
-#> os macOS Ventura 13.0
-#> system aarch64, darwin20
-#> ui X11
-#> language (EN)
-#> collate en_US.UTF-8
-#> ctype en_US.UTF-8
-#> tz Europe/Berlin
-#> date 2022-11-12
-#> pandoc 2.19.2 @ /Applications/RStudio.app/Contents/MacOS/quarto/bin/tools/ (via rmarkdown)
-#>
-#> ─ Packages ───────────────────────────────────────────────
-#> ! package * version date (UTC) lib source
-#> assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.2.0)
-#> backports 1.4.1 2021-12-13 [1] CRAN (R 4.2.0)
-#> base * 4.2.2 2022-10-31 [?] local
-#> bench * 1.1.2 2021-11-30 [1] CRAN (R 4.2.0)
-#> bookdown 0.30 2022-11-09 [1] CRAN (R 4.2.2)
-#> broom 1.0.1 2022-08-29 [1] CRAN (R 4.2.0)
-#> bslib 0.4.1 2022-11-02 [1] CRAN (R 4.2.2)
-#> cachem 1.0.6 2021-08-19 [1] CRAN (R 4.2.0)
-#> cellranger 1.1.0 2016-07-27 [1] CRAN (R 4.2.0)
-#> cli 3.4.1 2022-09-23 [1] CRAN (R 4.2.0)
-#> colorspace 2.0-3 2022-02-21 [1] CRAN (R 4.2.0)
-#> P compiler 4.2.2 2022-10-31 [1] local
-#> crayon 1.5.2 2022-09-29 [1] CRAN (R 4.2.1)
-#> P datasets * 4.2.2 2022-10-31 [1] local
-#> DBI 1.1.3.9002 2022-10-17 [1] Github (r-dbi/DBI@2aec388)
-#> dbplyr 2.2.1 2022-06-27 [1] CRAN (R 4.2.0)
-#> digest 0.6.30 2022-10-18 [1] CRAN (R 4.2.1)
-#> downlit 0.4.2 2022-07-05 [1] CRAN (R 4.2.1)
-#> dplyr * 1.0.10 2022-09-01 [1] CRAN (R 4.2.1)
-#> ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.2.0)
-#> evaluate 0.18 2022-11-07 [1] CRAN (R 4.2.2)
-#> fansi 1.0.3 2022-03-24 [1] CRAN (R 4.2.0)
-#> farver 2.1.1 2022-07-06 [1] CRAN (R 4.2.1)
-#> fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.2.0)
-#> forcats * 0.5.2 2022-08-19 [1] CRAN (R 4.2.1)
-#> fs 1.5.2 2021-12-08 [1] CRAN (R 4.2.0)
-#> gargle 1.2.1 2022-09-08 [1] CRAN (R 4.2.1)
-#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.2.1)
-#> ggplot2 * 3.4.0 2022-11-04 [1] CRAN (R 4.2.2)
-#> glue 1.6.2 2022-02-24 [1] CRAN (R 4.2.0)
-#> googledrive 2.0.0 2021-07-08 [1] CRAN (R 4.2.0)
-#> googlesheets4 1.0.1 2022-08-13 [1] CRAN (R 4.2.0)
-#> P graphics * 4.2.2 2022-10-31 [1] local
-#> P grDevices * 4.2.2 2022-10-31 [1] local
-#> P grid 4.2.2 2022-10-31 [1] local
-#> gtable 0.3.1 2022-09-01 [1] CRAN (R 4.2.1)
-#> haven 2.5.1 2022-08-22 [1] CRAN (R 4.2.0)
-#> highr 0.9 2021-04-16 [1] CRAN (R 4.2.0)
-#> hms 1.1.2 2022-08-19 [1] CRAN (R 4.2.0)
-#> htmltools 0.5.3 2022-07-18 [1] CRAN (R 4.2.1)
-#> httr 1.4.4 2022-08-17 [1] CRAN (R 4.2.0)
-#> jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.2.0)
-#> jsonlite 1.8.3 2022-10-21 [1] CRAN (R 4.2.1)
-#> knitr 1.40 2022-08-24 [1] CRAN (R 4.2.1)
-#> labeling 0.4.2 2020-10-20 [1] CRAN (R 4.2.0)
-#> lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.2.1)
-#> lobstr * 1.1.2 2022-06-22 [1] CRAN (R 4.2.0)
-#> lubridate 1.9.0 2022-11-06 [1] CRAN (R 4.2.2)
-#> magrittr * 2.0.3 2022-03-30 [1] CRAN (R 4.2.0)
-#> memoise 2.0.1 2021-11-26 [1] CRAN (R 4.2.0)
-#> P methods * 4.2.2 2022-10-31 [1] local
-#> modelr 0.1.10 2022-11-11 [1] CRAN (R 4.2.2)
-#> munsell 0.5.0 2018-06-12 [1] CRAN (R 4.2.0)
-#> pillar 1.8.1 2022-08-19 [1] CRAN (R 4.2.1)
-#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.2.0)
-#> prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.2.0)
-#> profmem 0.6.0 2020-12-13 [1] CRAN (R 4.2.0)
-#> purrr * 0.3.5 2022-10-06 [1] CRAN (R 4.2.1)
-#> R6 2.5.1.9000 2022-10-27 [1] local
-#> readr * 2.1.3 2022-10-01 [1] CRAN (R 4.2.1)
-#> readxl 1.4.1 2022-08-17 [1] CRAN (R 4.2.0)
-#> reprex 2.0.2 2022-08-17 [1] CRAN (R 4.2.1)
-#> rlang 1.0.6 2022-09-24 [1] CRAN (R 4.2.1)
-#> rmarkdown 2.18 2022-11-09 [1] CRAN (R 4.2.2)
-#> rstudioapi 0.14 2022-08-22 [1] CRAN (R 4.2.1)
-#> rvest 1.0.3 2022-08-19 [1] CRAN (R 4.2.1)
-#> sass 0.4.2 2022-07-16 [1] CRAN (R 4.2.1)
-#> scales 1.2.1 2022-08-20 [1] CRAN (R 4.2.1)
-#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.2.0)
-#> P stats * 4.2.2 2022-10-31 [1] local
-#> stringi 1.7.8 2022-07-11 [1] CRAN (R 4.2.1)
-#> stringr * 1.4.1 2022-08-20 [1] CRAN (R 4.2.1)
-#> tibble * 3.1.8.9002 2022-10-16 [1] local
-#> tidyr * 1.2.1 2022-09-08 [1] CRAN (R 4.2.1)
-#> tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.2.1)
-#> tidyverse * 1.3.2 2022-07-18 [1] CRAN (R 4.2.0)
-#> timechange 0.1.1 2022-11-04 [1] CRAN (R 4.2.2)
-#> P tools 4.2.2 2022-10-31 [1] local
-#> tzdb 0.3.0 2022-03-28 [1] CRAN (R 4.2.0)
-#> utf8 1.2.2 2021-07-24 [1] CRAN (R 4.2.0)
-#> P utils * 4.2.2 2022-10-31 [1] local
-#> vctrs 0.5.0 2022-10-22 [1] CRAN (R 4.2.1)
-#> withr 2.5.0 2022-03-03 [1] CRAN (R 4.2.0)
-#> xfun 0.34 2022-10-18 [1] CRAN (R 4.2.1)
-#> xml2 1.3.3.9000 2022-10-10 [1] local
-#> yaml 2.3.6 2022-10-18 [1] CRAN (R 4.2.1)
-#>
-#> [1] /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library
-#>
-#> P ── Loaded and on-disk path mismatch.
-#>
-#> ──────────────────────────────────────────────────────────
-```
diff --git a/_book/Names-values_files/figure-html/Names-values-31-1.png b/_book/Names-values_files/figure-html/Names-values-31-1.png
deleted file mode 100644
index 6210e6f4..00000000
Binary files a/_book/Names-values_files/figure-html/Names-values-31-1.png and /dev/null differ
diff --git a/_book/OO-tradeoffs.md b/_book/OO-tradeoffs.md
deleted file mode 100644
index afc23c3c..00000000
--- a/_book/OO-tradeoffs.md
+++ /dev/null
@@ -1,3 +0,0 @@
-# Trade-offs
-
-No exercises.
diff --git a/_book/Perf-improve.md b/_book/Perf-improve.md
deleted file mode 100644
index c9dc4ff3..00000000
--- a/_book/Perf-improve.md
+++ /dev/null
@@ -1,606 +0,0 @@
-# Improving performance
-
-
-
-Attaching the needed libraries:
-
-
-```r
-library(ggplot2)
-library(dplyr)
-library(purrr)
-```
-
-## Exercises 24.3.1
-
-**Q1.** What are faster alternatives to `lm()`? Which are specifically designed to work with larger datasets?
-
-**A1.** Faster alternatives to `lm()` can be found by visiting [CRAN Task View: High-Performance and Parallel Computing with R](https://cran.r-project.org/web/views/HighPerformanceComputing.html) page.
-
-Here are some of the available options:
-
-- `speedglm::speedlm()` (for large datasets)
-
-- `biglm::biglm()` (specifically designed for data too large to fit in memory)
-
-- `RcppEigen::fastLm()` (using the `Eigen` linear algebra library)
-
-High performances can be obtained with these packages especially if R is linked against an optimized BLAS, such as ATLAS. You can check this information using `sessionInfo()`:
-
-
-```r
-sessInfo <- sessionInfo()
-sessInfo$matprod
-#> [1] "default"
-sessInfo$LAPACK
-#> [1] "/Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/lib/libRlapack.dylib"
-```
-
-Comparing performance of different alternatives:
-
-
-```r
-library(gapminder)
-
-# having a look at the data
-glimpse(gapminder)
-#> Rows: 1,704
-#> Columns: 6
-#> $ country  -
-**Q3.** How can you use `crossprod()` to compute a weighted sum? How much faster is it than the naive `sum(x * w)`?
-
-**A3.** Both of these functions provide a way to compute a weighted sum:
-
-
-```r
-x <- c(1:6, 2, 3)
-w <- rnorm(length(x))
-
-crossprod(x, w)[[1]]
-#> [1] 15.94691
-sum(x * w)[[1]]
-#> [1] 15.94691
-```
-
-But benchmarking their performance reveals that the latter is significantly faster than the former!
-
-
-```r
-bench::mark(
- crossprod(x, w)[[1]],
- sum(x * w)[[1]],
- iterations = 1e6
-)[1:5]
-#> # A tibble: 2 × 5
-#> expression min median `itr/sec` mem_alloc
-#>
-
-**Q3.** How can you use `crossprod()` to compute a weighted sum? How much faster is it than the naive `sum(x * w)`?
-
-**A3.** Both of these functions provide a way to compute a weighted sum:
-
-
-```r
-x <- c(1:6, 2, 3)
-w <- rnorm(length(x))
-
-crossprod(x, w)[[1]]
-#> [1] 15.94691
-sum(x * w)[[1]]
-#> [1] 15.94691
-```
-
-But benchmarking their performance reveals that the latter is significantly faster than the former!
-
-
-```r
-bench::mark(
- crossprod(x, w)[[1]],
- sum(x * w)[[1]],
- iterations = 1e6
-)[1:5]
-#> # A tibble: 2 × 5
-#> expression min median `itr/sec` mem_alloc
-#>  -
----
-
-## Quoting (Exercises 19.3.6)
-
----
-
-**Q1.** How is `expr()` implemented? Look at its source code.
-
-**A1.** Looking at the source code, we can see that `expr()` is a simple wrapper around `enexpr()`, and captures and returns the user-entered expressions:
-
-
-```r
-rlang::expr
-#> function (expr)
-#> {
-#> enexpr(expr)
-#> }
-#>
-
----
-
-## Quoting (Exercises 19.3.6)
-
----
-
-**Q1.** How is `expr()` implemented? Look at its source code.
-
-**A1.** Looking at the source code, we can see that `expr()` is a simple wrapper around `enexpr()`, and captures and returns the user-entered expressions:
-
-
-```r
-rlang::expr
-#> function (expr)
-#> {
-#> enexpr(expr)
-#> }
-#>