Authors: Maxime Gillot Baptiste Baquero Antonio Ruellas Marcela Gurgel Najla Al Turkestani Elizabeth Biggs Marilia Yatabe Jonas Bianchi Lucia Cevidanes Juan Carlos Prieto
We propose a novel approach that reformulates landmark detection as a classification problem through a virtual agent placed inside a 3D Cone-Beam Computed Tomography (CBCT) scan. This agent is trained to navigate in a multi-scale volumetric space to reach the estimated landmark position.
Scripts for Automatic Landmark Identification in CBCT
python 3.8.8 with the anaconda environment "ALI_environment.yml":
Main librairies:
monai==0.7.0
torch==1.10.1
itk==5.2.1
numpy==1.20.1
simpleitk==2.1.1
You can get the ALI CBCT docker image by running the folowing command lines.
Informations
- A test scan "MG_scan_test.nii.gz" is provided in the Data folder of the ALI_CBCT repositorie.
- If the prediction with the GPU is not working, make sure you installed the NVIDIA Container Toolkit : https://docs.nvidia.com/datacenter/cloud-native/container-toolkit/install-guide.html#docker
Building using the DockerFile
From the DockerFile directory:
docker pull dcbia/alicbct:latest
From the DockerFile directory:
docker build -t alicbct .
Automatic landmark identification Running on CPU
docker run --rm --shm-size=5gb -v <Folder_with_the_scans_to_segment>:/app/data/scans -v <Folder_with_the_models_to_use>:/app/data/models alicbct:latest python3 /app/ALI_CBCT/predict_landmarks.py
Running on GPU
docker run --rm --shm-size=5gb --gpus all -v <Folder_with_the_scans_to_segment>:/app/data/scans -v <Folder_with_the_models_to_use>:/app/data/models alicbct:latest python3 /app/ALI_CBCT/predict_landmarks.py
options/arguments
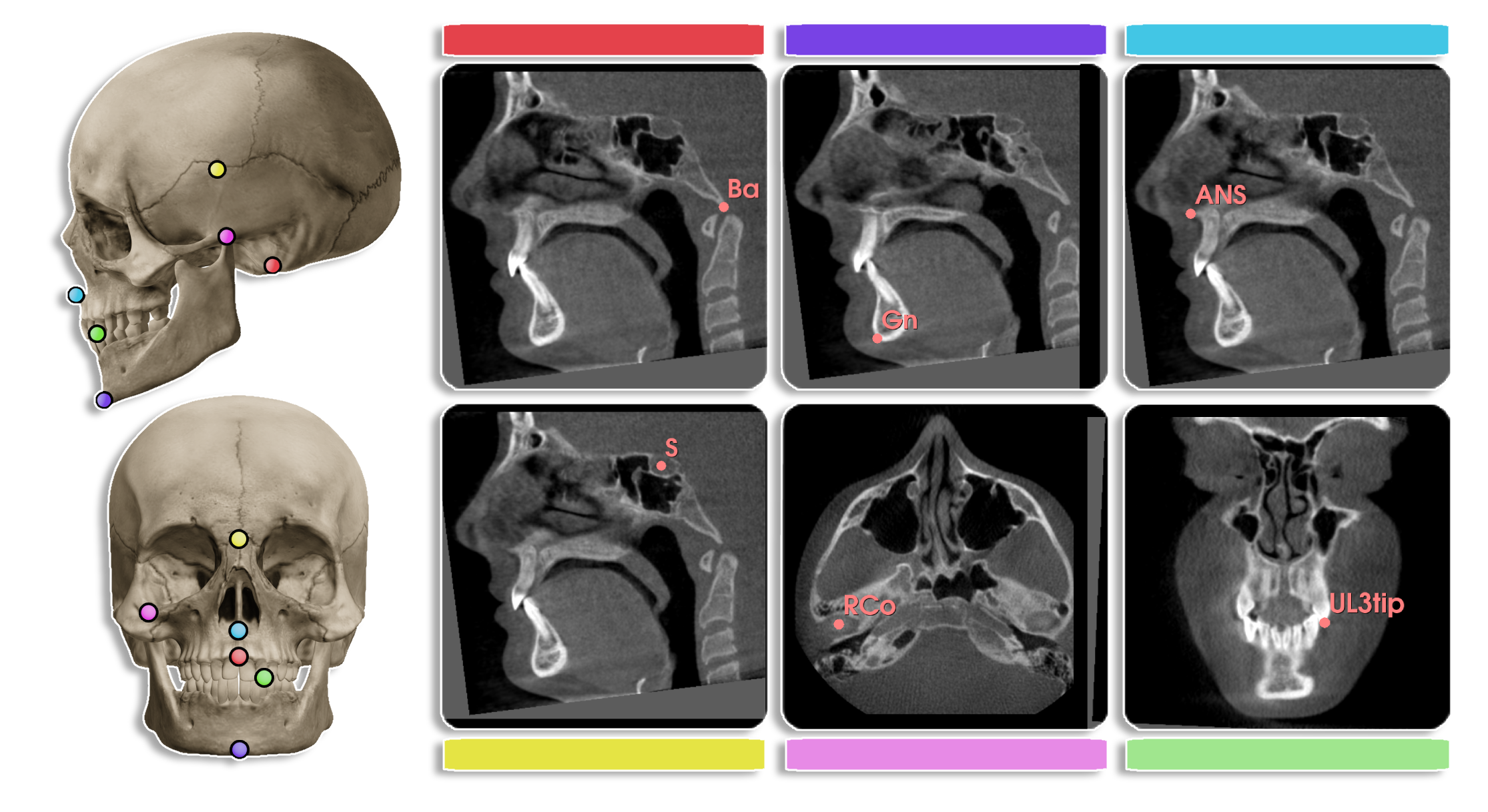
- By default, only Ba and S landmarks are selected.
To choose which structure to segment, you can use the following arguments:
-lm Ba S
Pre-process
To run the preprocess to prepare the files and set them at the wanted spacing for the training:
For the Upper, Lower and Cranial base landmarks
python3 init_training_data_ULCB.py -i "path of the input folder with the scans and the fiducial list" -o "path of the output folder"
For the canine impaction landmarks
python3 init_training_data_Canine.py -i "path of the input folder with the scans and the fiducial list" -o "path of the output folder"
By default the spacing is set at 1 and 0.3 but we can change and add other spacing with the argument :
-sp x.xx x.xx
A contrast adjustment is also applied but can be turned off with
-ch False
#Images