-
Notifications
You must be signed in to change notification settings - Fork 9
Exercise D: Plotting Your Results with PyROOT
We're not quite done yet because we don't just publish .root files for other researchers to look at. We publish plots, graphical representations of histograms. A plot might display only a certain bin range, display multiple histograms together, or split a multi-dimensional histogram into multiple 1D histograms.
We separate plotting from histogram filling in MINERvA analyses because plotting is usually a lot faster than the event loop. Imagine rerunning runEventLoop with full systematics just to change the units on your x axis from "GeV" to "GeV/c"! We produced histograms yesterday, and we've turned them into quick-and-dirty plots during the closure test and warping studies. But for publication-quality plots, interactive ROOT sessions aren't usually enough. You might have to worry about:
- Which files you compare
- What exactly is in the axis labels
- Displaying related plots together in a regular grid
- Breaking our systematic uncertainties down into the best possible categories
- Whether the line colors you use are distinguishable to color-blind researchers
We can write plotting scripts to make our solutions to these problems reproducible and easy to update. ROOT's interpreter can actually read a series of plotting commands from a file. But we're going to use python to make our plotting macros shorter and easier to debug. MINERvAns often choose scripts for plotting over full programs because scripting language programs are easier to update and don't need robust user interfaces.
ROOT provides a lot of tools for plotting histograms. The vast majority of them can be accessed from the TH1 base class that all histograms (even PlotUtils::MnvH1D) derive from. But TH1 itself is very complex, so I'll break down some commonly used tools.
We might want to perform mathematical histograms on histograms before plotting them. TH1 provides the interface that allow us to plot ratios of histograms, area normalize them, and subtract backgrounds. Some common histogram operations are:
-
TH1::Scale(): multiply each bin by the same constant. If that constant is theTH1::GetIntegral(), then the scaled histogram is said to be area-normalized. -
TH1::Divide(): take the ratio of two histograms. We usually useTH1::Divide(TH1*, TH1*, /*...*/), which replaces the contents of the first histogram with the ratio, because it's the only interface that propagates systematics betweenMnvH1Ds. -
TH1::Add(): add corresponding bins from two histograms that have the same axes. Notice the multiplicative factor in the second parameter. We use -1 for the scale factor to subtract background histograms from the data inExtractCrossSection.
TH1::Draw() (really TObject::Draw()) takes a string as its only argument to control options as diverse as not redrawing the current TCanvas and drawing a 2D histogram as a contour plot instead of a heatmap. These so-called "draw options" are actually documented in a separate class, THistPainter. I'll highlight a few I use often:
- SAME: Draw a histogram on the same
TCanvasas already-drawn histogram. CallingTH1::Draw()with no arguments erases an existingTCanvasand redraws it with just the new histogram. - COLZ: Draw a 2D histogram as a heatmap. The default
TH2::Draw()behavior is kind of hard to interpret in most cases. We use this often withTEXTto represent migration and covariance matrices. - E3: Draw error bars as an error envelope instead. I usually use this with
SetFillColorAlpha()to make sure the error envelope is transparent. - HIST: Don't draw error bars at all. Sometimes, the default statistical uncertainties ROOT calculates are distracting or misleading.
You'll notice in the example that I also interact with the axes of a plot, via TAxis, the "window" the histogram is plotted in, via TCanvas, the TLegend, and a global style variable, gStyle.
A MINERvA-specific package called PlotUtils associates systematic Universes with "central value", or unmodified, histograms. So, we have to provide plotting functionality for summarizing systematic uncertainties too. The PlotUtils "histogram with systematics" class, MnvH1D, broadcasts operations like Add() and Scale() to each Universe's histogram. The error bands on an MnvH1D are statistical-only by default though. To get full error bands, you have to GetCVHistoWithSystematics(). MnvPlotter has lots of useful functions like PlotErrorSummary() for making sense of Universe histograms as systematic uncertainties.
We are in the process of releasing PlotUtils to the public via github. Check back on this project in a couple of months.
ROOT ships with "python bindings" that make c++ objects available as python objects in a module named ROOT. So, a TH1D histogram could be created like: hist = ROOT.TH1D("name", "title", nBins, min, max). Python works well with ROOT because they use the same kind of memory model. ROOT often returns opaque pointers to a universal base class called TObject, and pyROOT lets us use the full list of functions available to the underlying object. Make ROOT's python bindings available in a python script by importing it like a module: import ROOT. ROOT also comes with machinery to generate python bindings for user-defined classes when it generates a "dictionary" for c++ code. We take advantage of this in PlotUtils, so you can also from ROOT import PlotUtils.
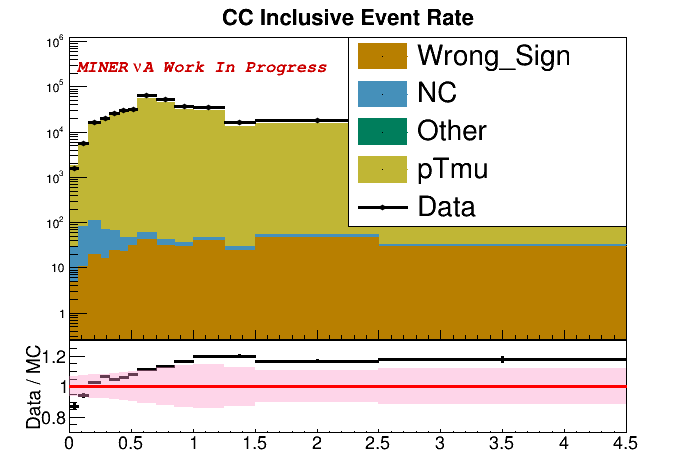
- Write a python script that compares the cross section from data with the cross section GENIEXSecExtract predicted. Start with just opening the right files and
Draw()ing both histograms at once. Then try to produce a second data/MC ratio plot with an error envelope around the MC line and points for the data. As a final challenge, try merging both plots into one plot on the sameTCanvas. - In Amy's 2D Inclusive cross section Wine and Cheese presentation, one of her plots broke down the predicted event rate by which GENIE process produced each neutrino interaction. Using the background histograms as an example, add a
Categorized<>torunEventLoopthat breaks the selected events down byGetInteractionType(). If you follow Amy's work closely, you'll notice that she breaks GENIE's DIS category into "Soft DIS" and "True DIS" categories based on kinematics. Finally, write a python script to plot the total predicted selected event rate along with the individual GENIE categories and data points.
- When I run
backgroundStack.py, I get messages like:
Error in <TInterpreter::TCling::AutoLoad>: failure loading library libG__PlotUtilsDict.so for PlotUtils::MnvH1D
Error in <TInterpreter::TCling::AutoLoad>: failure loading library libG__PlotUtilsDict.so for PlotUtils::MnvH1D
TClass::Init:0: RuntimeWarning: no dictionary for class PlotUtils::MnvH1D is available
Error in <TInterpreter::TCling::AutoLoad>: failure loading library libG__PlotUtilsDict.so for PlotUtils::MnvH1D
Error in <TInterpreter::TCling::AutoLoad>: failure loading library libG__PlotUtilsDict.so for PlotUtils::MnvVertErrorBand
Error in <TInterpreter::TCling::AutoLoad>: failure loading library libG__PlotUtilsDict.so for PlotUtils::MnvVertErrorBand
Solution: make sure you run in a directory with a .rootlogon.C file like the one in Prerequisites
- I get a message about ROOT not being a module:
from ROOT import *
ImportError: No module named ROOT
Solution: Copy backgroundStack.py to your working directory and run it like this instead: python3 backgroundStack.py. The problem is that your ROOT installation was built against python3, but your system python command points to python 2. I ran out of time to juggle this requirements across platforms.