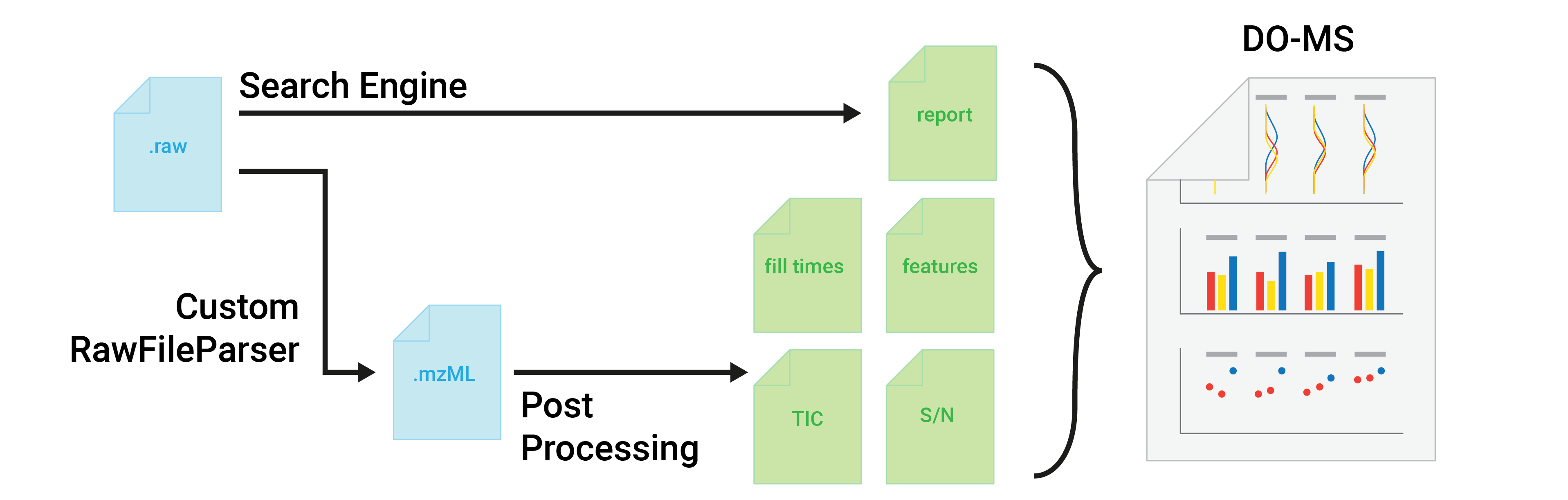
Data-Driven Optimization of Mass Spectrometry Methods
Please read our detailed getting started guides:
- Getting started with DIA preprocessing
- Getting started with DIA reports
- Getting started with DDA reports
This application has been tested on R >= 3.5.0, OSX 10.14 / Windows 7/8/10/11. R can be downloaded from the main R Project page or downloaded with the RStudio Application. All modules are maintained for MaxQuant >= 1.6.0.16 and DIA-NN > 1.8.1.
The application suffers from visual glitches when displayed on unsupported older browsers (such as IE9 commonly packaged with RStudio on Windows). Please use IE >= 11, Firefox, or Chrome for the best user experience.
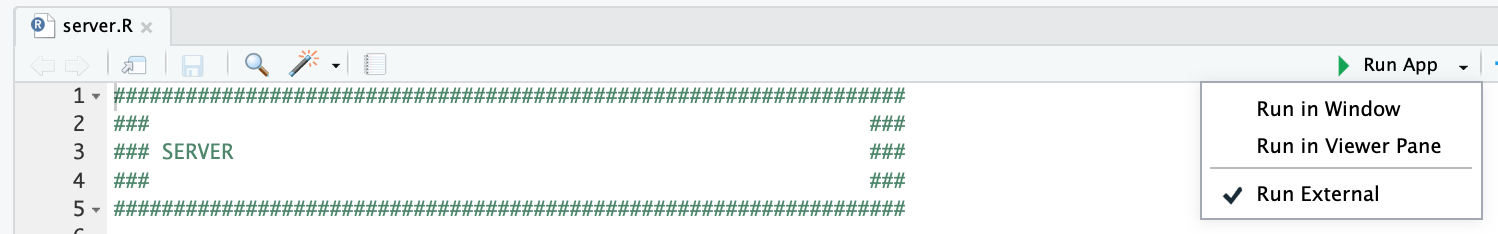
The easiest way to run the app is directly through RStudio, by opening the DO-MS.Rproj Rproject file
and clicking the "Run App" button at the top of the application, after opening the server.R file. We recommend checking the "Run External" option to open the application in your default browser instead of the RStudio Viewer.
You can also start the application by running the start_server.R script.
DO-MS is designed to be easily user-customizable for in-house proteomics workflows. Please see Building Your Own Modules for more details.
Please see Hosting as a Server for more details.
This application is currently maintained for (MaxQuant)[https://www.nature.com/articles/nbt.1511] >= 1.6.0.16 and (DIA-NN)[https://www.nature.com/articles/s41592-019-0638-x] >= 1.8. Adapting to other search engines is possible but not provided out-of-the-box. Please see Integrating Other Search Engines for more details.
While the base library of modules are based around bottom-up proteomics by LC-MS/MS, this project is fundamentally compatible with any delimited text files (CSV, TSV, etc). These implementations will require some programming work, but once it is done DO-MS gives you a extensible framework that can be used over-and-over again to generate shareable reports. See Integrating Other Search Engines for more details
The manuscript for this tool is published at the Journal of Proteome Research: https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00039 The manuscript for the extended version 2.0 can be found on bioArxiv: https://www.biorxiv.org/content/10.1101/2023.02.02.526809v1
Contact the authors by email: nslavov{at}northeastern.edu.
DO-MS is distributed by an MIT license.
Please feel free to contribute to this project by opening an issue or pull request in the GitHub repository.
For any bugs, questions, or feature requests, please use the GitHub issue system to contact the developers.