Reminder¶
Where are we? Where are we going?
+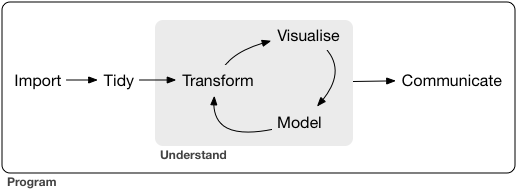 +
+source: R for Data Science by Grolemund & Wickham
+Classification¶
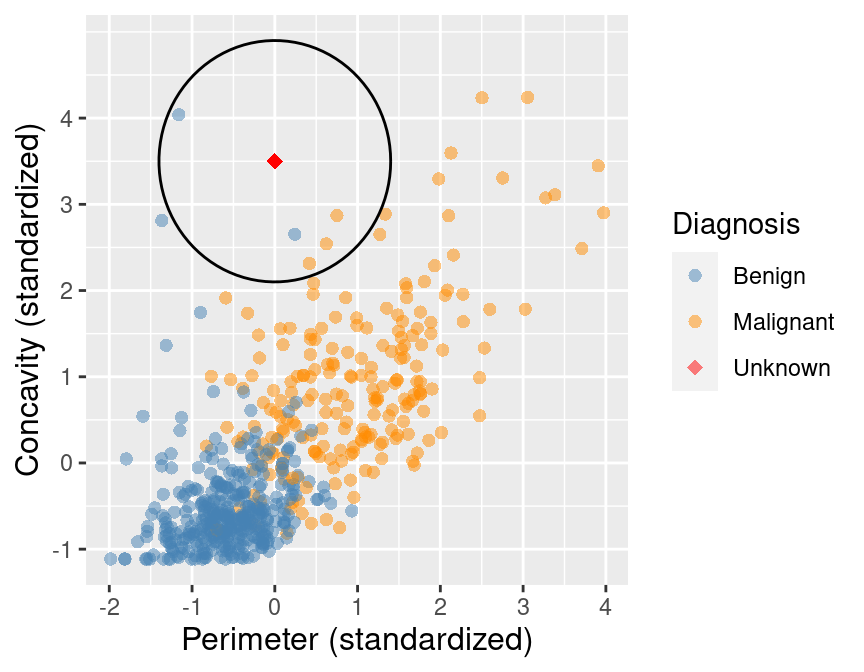
Suppose we have past data of cancer tumour cell diagnosis labelled "benign" and "malignant". Do you think a new cell with Concavity = 3.3 and Perimeter = 0.2 would be malignant? How did you decide?
+
What kind of data analysis question is this?¶
Descriptive, exploratory, predictive, inferential, causal, or mechanistic?
+K-nearest neighbours classification¶
Predict the label / class for a new observation using the K closest points from our dataset.
+-
+
- Compute the distance between the new observation and each observation in our training set
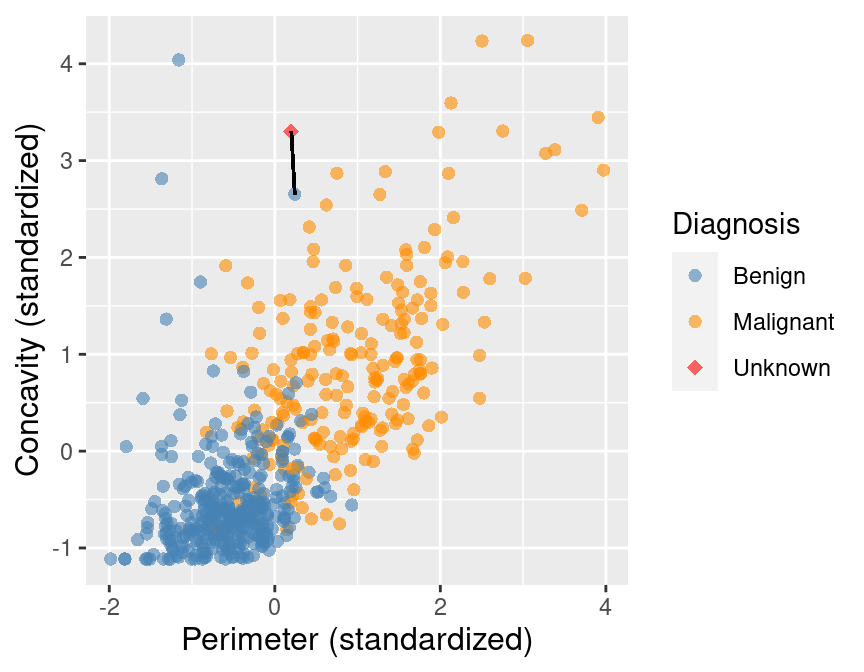
K-nearest neighbours classification¶
Predict the label / class for a new observation using the K closest points from our dataset.
+-
+
- Sort the data in ascending order according to the distances +
- Choose the top K rows as "neighbours" +
## # A tibble: 5 x 5
+## ID Perimeter Concavity Class dist_from_new
+## <dbl> <dbl> <dbl> <fct> <dbl>
+## 1 86409 0.241 2.65 B 0.881
+## 2 887181 0.750 2.87 M 0.980
+## 3 899667 0.623 2.54 M 1.14
+## 4 907914 0.417 2.31 M 1.26
+## 5 8710441 -1.16 4.04 B 1.28
+We can go beyond 2 predictors¶
For two observations $u, v$, each with $m$ variables (columns) labelled $1, \dots, m$,
+
+
Aside from that, it's the same algorithm!
+Standardizing Data¶


Nonstandardized Data vs Standardized Data¶
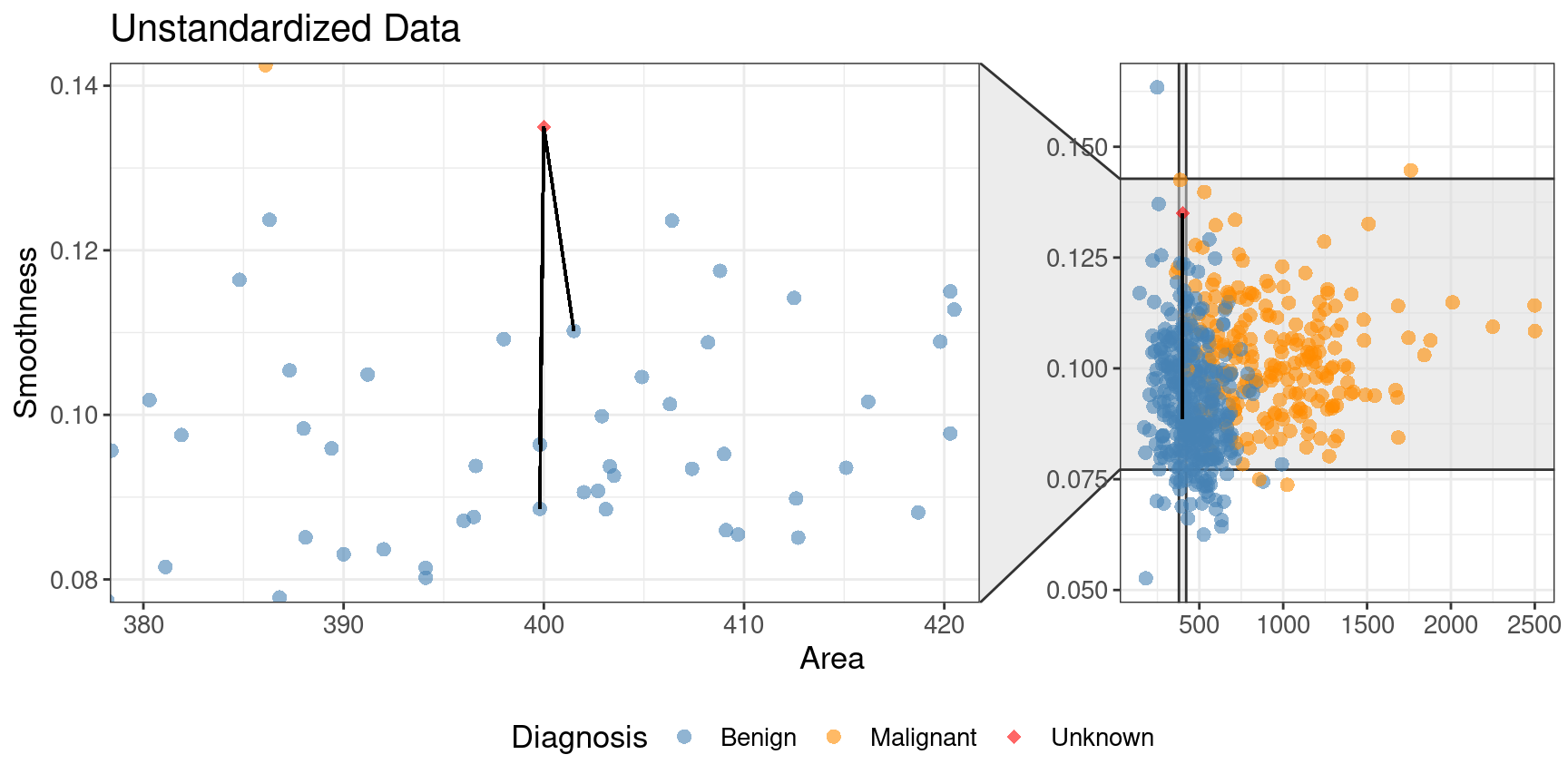
What if one variable is much larger than the other?
+Standardize: shift and scale so that the average is 0 and the standard deviation is 1.
++
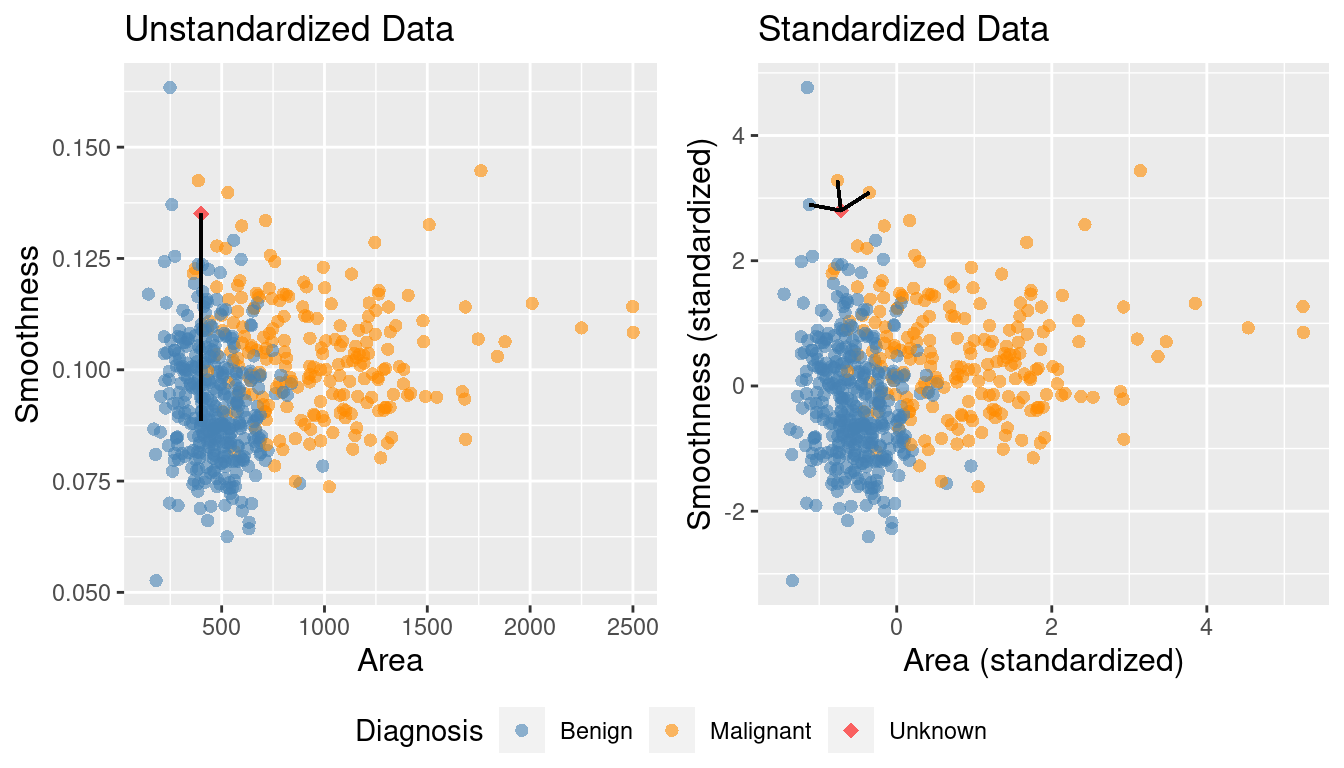 +
+Introduction to the tidymodels package in R¶
tidymodels is a collection of packages and handles computing distances, standardization, balancing, and prediction for us!
 +
+Introduction to the tidymodels package in R¶
-
+
- Load the libraries and data we need (new:
tidymodels)
+
library(tidyverse)
+library(tidymodels)
+options(repr.matrix.max.rows = 6)
+
+# Data on cancer tumors
+tumors <- read_csv("data/clean-wdbc.data.csv") |>
+ mutate(Class = as_factor(Class))
+Introduction to the tidymodels package in R¶
0b. Inspect the data
+tumors
+Introduction to the tidymodels package in R¶
In tidymodels, the recipes package is named after cooking terms.
1. Make a recipe to specify the predictors/response and preprocess the data¶
-
+
+recipe(): Main argument in the formula.Arguments:
+-
+
- formula +
- data +
+
+prep()&bake(): you can alsoprepandbakea recipe to see what the preprocessing does!
+
-
+
- visit https://recipes.tidymodels.org/reference/index.html to see all the preprocessing steps +
tumors |> head(2)
+
+# Standardize the cancer data
+Introduction to the tidymodels package in R¶
2. Build a model specification (model_spec) to specify the model and training algorithm¶
-
+
model type: kind of model you want to fit
+
+arguments: model parameter values
+
+engine: underlying package the model should come from
+
+mode: type of prediction (some packages can do both classification and regression)
+
+
# Build a model specification using nearest_neighbor
+# Create a workflow
+new_obs <- tibble(Perimeter = 0.2, Concavity = 3.3)
+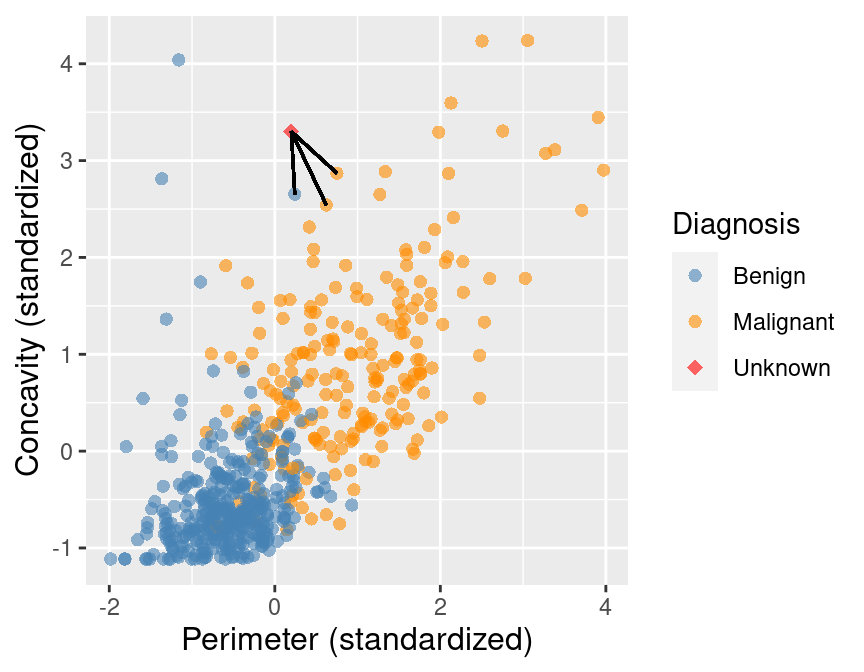 +
+What did we learn today?¶
-
+
+
+
+
+ +
+ +
+ +
+ +
+