-
Notifications
You must be signed in to change notification settings - Fork 0
/
index.Rmd
498 lines (339 loc) · 9.46 KB
/
index.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
---
title: "Bioreach"
subtitle: "Mission opportunity 2022"
author:
- "Bertrand Bhikarry"
# - "Garrick Aden-Buie"
date: '`r Sys.Date()`'
output:
xaringan::moon_reader:
css: xaringan-themer.css
nature:
slideNumberFormat: "%current%"
highlightStyle: github
highlightLines: true
ratio: 4:3
countIncrementalSlides: true
# includes:
# after_body: timer.html
---
```{r setup, include=FALSE}
options(htmltools.dir.version = FALSE)
knitr::opts_chunk$set(
fig.width=9, fig.height=3.5, fig.retina=3,
out.width = "100%",
cache = FALSE,
echo = TRUE,
message = FALSE,
warning = FALSE,
hiline = TRUE
)
```
```{r xaringan-themer, include=FALSE, warning=FALSE}
library(xaringanthemer)
#style_duo_accent(
# primary_color = "#035AA6",
# secondary_color = "#FF961C",
# inverse_header_color = "#FFFFFF"
#)
# new style
style_mono_accent(
base_color = "#1c5253",
header_font_google = google_font("Josefin Sans"),
text_font_google = google_font("Montserrat", "300", "300i"),
code_font_google = google_font("Fira Mono")
)
```
```{r, include=FALSE}
library(countdown)
```
```{r xaringan-logo, echo=FALSE}
xaringanExtra::use_logo(
image_url = "https://drive.google.com/uc?export=view&id=11GveNpJ7VdDl4t1pUGszDuM10P-KCMaB"
)
```
```{r, echo=FALSE}
xaringanExtra::use_webcam(width = 300, height = 225)
```
```{r xaringan-panelset, echo=FALSE}
xaringanExtra::use_panelset()
```
```{r xaringanExtra, echo=FALSE}
xaringanExtra::use_xaringan_extra(c("tile_view", "animate_css", "tachyons"))
```
## Biodiversity Conservation and Agroecological Land Restoration in Productive Landscapes of T&T
**Duration** 4 year (Pilot)
**Funding** USD 22,454,792 (GEF/GoRTT)
**Executed by**
1. Environmental Management Auth (EMA)
1. Nat'l Agricultural Marketing and Development Co (NAMDEVCO)
**Implementing agency** Food and Agriculture Org (UN/FAO)
---
## Needed why?
### Big picture
1. To protect biodiversity in 'productive' spaces
1. To protect arable land from poor land use and invasive species
1. To enhance nature's services focussing on forests
1. Develop model green value chains in the agri sector
1. Develop sustainable agro-eco-forestry livelihoods
---
## Bioreach has 4 components
.panelset[
.panel[.panel-name[Component 1]
#### Biodiversity sensitive and supportive land-use planning mechanisms.
```{r, echo=FALSE, out.width="90%",fig.align='center', fig.cap="A stakeholder session"}
knitr::include_graphics("https://drive.google.com/uc?export=view&id=1VBklp7M_bYrmew2Eo4w4zmWxgp1QzCHQ")
```
]
.panel[.panel-name[Component 2]
#### Forests and Agricultural landscape restoration and biodiversity protection thru Agroecology.
```{r, echo=FALSE, fig.align='center', out.width='40%', fig.cap='Increase agriculture but not at the expense of tree cover.'}
knitr::include_graphics("https://drive.google.com/uc?export=view&id=10DS97WWiWxT_ryvViO5SwoG2CIwpqXPc")
```
]
.panel[.panel-name[Component 3]
#### Enabling environment for green biodiversity-friendly value chains.
```{r, echo=FALSE, out.width='45%', fig.align='center'}
knitr::include_graphics("https://drive.google.com/uc?export=view&id=1TVaiqICZWi8aB7SVCYefVu0Drz834Zkx")
```
]
.panel[.panel-name[Component 4]
#### Knowledge management and monitoring.
<!--- a table --->
|**Outcome**|**Outputs**|
|:-----|:-----|
|Improved knowledge management on biodiversity and land degradation issues.|Produce and dissemine the info developed across T&T |
|Ongoing monitoring feeds into project as it happens|Ensure gender balance (*to be looked at annually*) |
]
]
???
See those Q marks? Thats a comment
---
## Summary. The Bioreach call is for
|**Target**|**Opportunity**|
|-----|-----|
|Active or wanna-be eco-agro foresters, agrofood-processors or nature conservationists| Training, property prepping (e.g soil testing), value chain identification and prepping, access to project lessons learnt|
???
Conservationists will be able to work with pawi (Trinidad) or bamboo if Tobago. As well as to collect data to pass to project leads.
---
## Typography
Text can be **bold**, _italic_, ~~strikethrough~~, or `inline code`.
[Link to another slide](#colors).
### Lorem Ipsum
Dolor imperdiet nostra sapien scelerisque praesent curae metus facilisis dignissim tortor.
Lacinia neque mollis nascetur neque urna velit bibendum.
Himenaeos suspendisse leo varius mus risus sagittis aliquet venenatis duis nec.
- Dolor cubilia nostra nunc sodales
- Consectetur aliquet mauris blandit
- Ipsum dis nec porttitor urna sed
---
name: colors
## Colors
.left-column[
Text color
[Link Color](#3)
**Bold Color**
_Italic Color_
`Inline Code`
]
.right-column[
Lorem ipsum dolor sit amet, [consectetur adipiscing elit (link)](#3),
sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
Erat nam at lectus urna.
Pellentesque elit ullamcorper **dignissim cras tincidunt (bold)** lobortis feugiat.
_Eros donec ac odio tempor_ orci dapibus ultrices.
Id porta nibh venenatis cras sed felis eget velit aliquet.
Aliquam id diam maecenas ultricies mi.
Enim sit amet
`code_color("inline")`
venenatis urna cursus eget nunc scelerisque viverra.
]
---
# Big Topic or Inverse Slides `#`
## Slide Headings `##`
### Sub-slide Headings `###`
#### Bold Call-Out `####`
This is a normal paragraph text. Only use header levels 1-4.
##### Possible, but not recommended `#####`
###### Definitely don't use h6 `######`
---
# Left-Column Headings
.left-column[
## First
## Second
## Third
]
.right-column[
Dolor quis aptent mus a dictum ultricies egestas.
Amet egestas neque tempor fermentum proin massa!
Dolor elementum fermentum pharetra lectus arcu pulvinar.
]
---
class: inverse center middle
# Topic Changing Interstitial
--
```
class: inverse center middle
```
---
layout: true
## Blocks
---
### Blockquote
> This is a blockquote following a header.
>
> When something is important enough, you do it even if the odds are not in your favor.
---
### Code Blocks
#### R Code
```{r eval=FALSE}
ggplot(gapminder) +
aes(x = gdpPercap, y = lifeExp, size = pop, color = country) +
geom_point() +
facet_wrap(~year)
```
#### JavaScript
```js
var fun = function lang(l) {
dateformat.i18n = require('./lang/' + l)
return true;
}
```
---
### More R Code
```{r eval=FALSE}
dplyr::starwars %>% dplyr::slice_sample(n = 4)
```
---
```{r message=TRUE, eval=requireNamespace("cli", quietly = TRUE)}
cli::cli_alert_success("It worked!")
```
--
```{r message=TRUE}
message("Just a friendly message")
```
--
```{r warning=TRUE}
warning("This could be bad...")
```
--
```{r error=TRUE}
stop("I hope you're sitting down for this")
```
---
layout: true
## Tables
---
exclude: `r if (requireNamespace("tibble", quietly=TRUE)) "false" else "true"`
```{r eval=requireNamespace("tibble", quietly=TRUE)}
tibble::as_tibble(mtcars)
```
---
```{r}
knitr::kable(head(mtcars), format = 'html')
```
---
exclude: `r if (requireNamespace("DT", quietly=TRUE)) "false" else "true"`
```{r eval=requireNamespace("DT", quietly=TRUE)}
DT::datatable(head(mtcars), fillContainer = FALSE, options = list(pageLength = 4))
```
---
layout: true
## Lists
---
.pull-left[
#### Here is an unordered list:
* Item foo
* Item bar
* Item baz
* Item zip
]
.pull-right[
#### And an ordered list:
1. Item one
1. Item two
1. Item three
1. Item four
]
---
### And a nested list:
- level 1 item
- level 2 item
- level 2 item
- level 3 item
- level 3 item
- level 1 item
- level 2 item
- level 2 item
- level 2 item
- level 1 item
- level 2 item
- level 2 item
- level 1 item
---
### Nesting an ol in ul in an ol
- level 1 item (ul)
1. level 2 item (ol)
1. level 2 item (ol)
- level 3 item (ul)
- level 3 item (ul)
- level 1 item (ul)
1. level 2 item (ol)
1. level 2 item (ol)
- level 3 item (ul)
- level 3 item (ul)
1. level 4 item (ol)
1. level 4 item (ol)
- level 3 item (ul)
- level 3 item (ul)
- level 1 item (ul)
---
layout: true
## Plots
---
```{r plot-example, eval=requireNamespace("ggplot2", quietly=TRUE)}
library(ggplot2)
(g <- ggplot(mpg) + aes(hwy, cty, color = class) + geom_point())
```
---
```{r plot-example-themed, eval=requireNamespace("showtext", quietly=TRUE) && requireNamespace("ggplot2", quietly=TRUE)}
g + xaringanthemer::theme_xaringan(text_font_size = 16, title_font_size = 18) +
ggtitle("A Plot About Cars")
```
.footnote[Requires `{showtext}`]
---
layout: false
## Square image
<center><img src="https://octodex.github.com/images/labtocat.png" alt="GithHub Octocat" height="400px" /></center>
.footnote[GitHub Octocat]
---
### Wide image
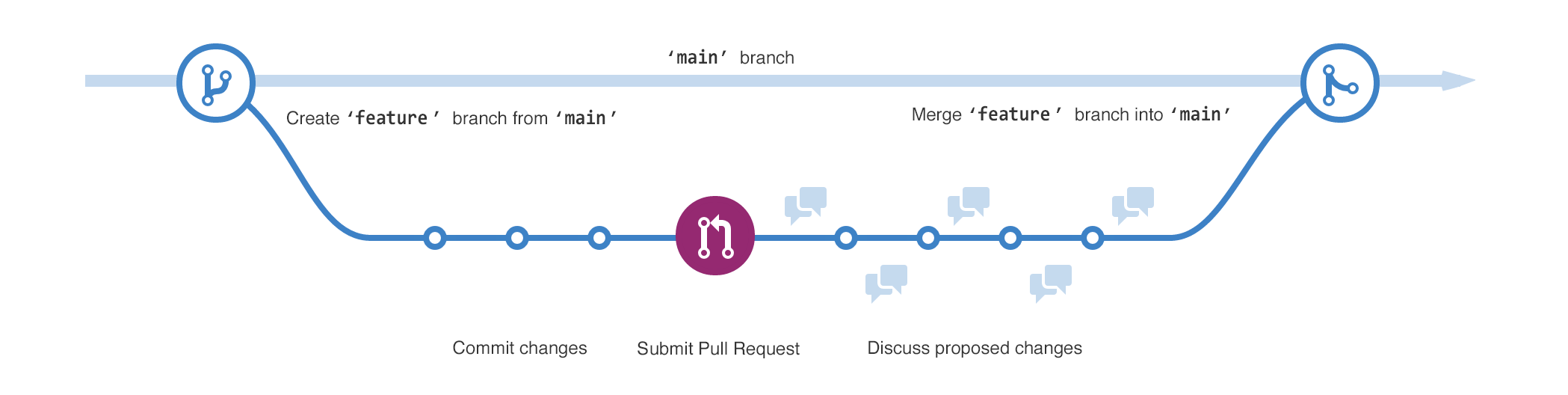
.footnote[Wide images scale to 100% slide width]
---
## Two images
.pull-left[

]
.pull-right[

]
---
### Definition lists can be used with HTML syntax.
<dl>
<dt>Name</dt>
<dd>Godzilla</dd>
<dt>Born</dt>
<dd>1952</dd>
<dt>Birthplace</dt>
<dd>Japan</dd>
<dt>Color</dt>
<dd>Green</dd>
</dl>
---
class: center, middle
# Thanks!
Slides created via the R packages:
[**xaringan**](https://github.com/yihui/xaringan)<br>
[gadenbuie/xaringanthemer](https://github.com/gadenbuie/xaringanthemer)
The chakra comes from [remark.js](https://remarkjs.com), [**knitr**](http://yihui.name/knitr), and [R Markdown](https://rmarkdown.rstudio.com).