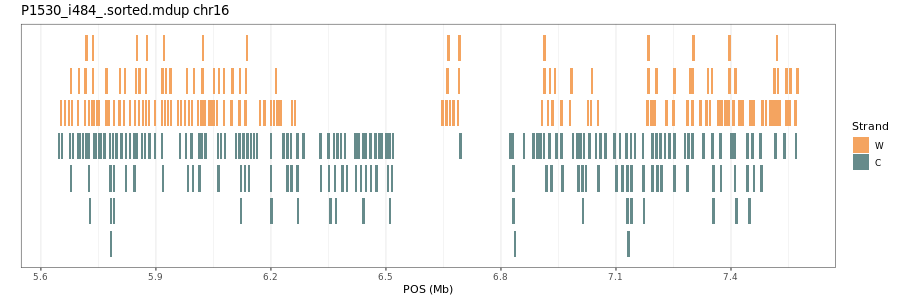
Workflow to plot the individual reads in a region of interest in a Strand-seq library. Written by Benedict March 2023, please let me know if you have any suggestions/issues!
Clone this repository like so:
git clone https://github.com/Sanders-Lab/strand-seq-read-plots
Then you can change directory into the created folder to run the scripts:
cd strand-seq-read-plots
For the shell script you will need samtools installed and in your environment (e.g. via conda).
For the R script you will require tidyverse for dplyr & ggplot2, which you can load with library(tidyverse) or install with `install.packages("tidyverse").
The first step is to execute 1_extract_reads.sh to extract the Watson and Crick reads:
bash 1_extract_reads.sh \
/fast/groups/ag_sanders/work/data/P1530/bam \
chr16:5643516-7570129 \
P1530_example_output.txt
Where the 1st command line argument is a directory containing the bam files of interest, the 2nd is the region of interest (format CHROM:Start-End), and the 3rd is a filepath for the output file.
Next you can plot the reads in your region of interest in R, by sourcing the plot_counts() function in 2_plot_reads.R.
Here is an example script to visualise a region of interest on chr16 on P1530_i484:
source('2_plot_reads.R')
library(tidyverse)
reads_df = read.table("P1530_example_output.txt.gz", header = T) %>%
filter(cell == "P1530_i484_.sorted.mdup")
png("P1530_chr16_example_plot.png", width = 900, height = 300)
plot_counts(input_df = reads_df)
dev.off()
The output is the graph below. Enjoy!