The following example may be launched interactively via any of the following:
This project was initially inspired by "BrainWeb: 20 Anatomical Models of 20 Normal Brains."
However there are a number of generally useful tools, image processing &
display functions included in this project. For example, this includes
volshow() for interactive comparison of multiple 3D volumes,
get_file() for caching data URLs, and register() for image
coregistration.
Download and Preprocessing for PET-MR Simulations
This notebook will not re-download/re-process files if they already exist.
- Output data
~/.brainweb/subject_*.npz: dtype(shape):float32(127, 344, 344)
- Raw data
source
~/.brainweb/subject_*.bin.gz: dtype(shape):uint16(362, 434, 362)
- Install
pip install brainweb
- Author: Casper da Costa-Luis <casper.dcl@physics.org>
- Date: 2017-2020
- Licence: MPLv2.0
from __future__ import print_function, division
%matplotlib notebook
import brainweb
from brainweb import volshow
import numpy as np
from os import path
from tqdm.auto import tqdm
import logging
logging.basicConfig(level=logging.INFO)# download
files = brainweb.get_files()
# read last file
data = brainweb.load_file(files[-1])
# show last subject
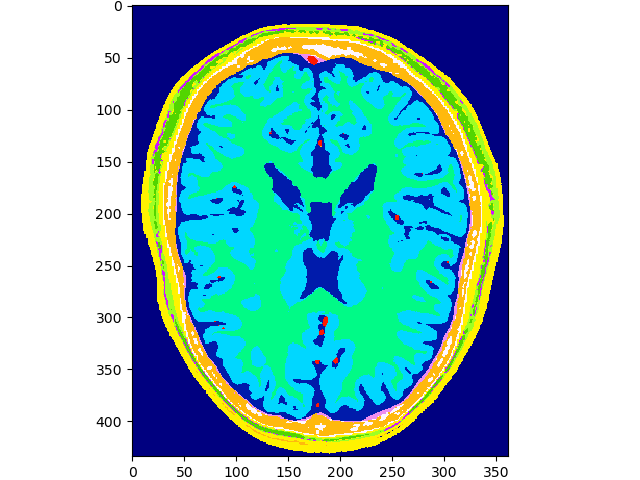
print(files[-1])
volshow(data, cmaps=['gist_ncar']);~/.brainweb/subject_54.bin.gz
Convert raw image data:
- Siemens Biograph mMR resolution (~2mm) & dimensions (127, 344, 344)
- PET/T1/T2/uMap intensities
- PET defaults to FDG intensity ratios; could use e.g. Amyloid instead
- randomised structure for PET/T1/T2
- t (1 + g [2 G_sigma(r) - 1]), where
- r = rand(127, 344, 344) in [0, 1),
- Gaussian smoothing sigma = 1,
- g = 1 for PET; 0.75 for MR, and
- t = the PET or MR piecewise constant phantom
- t (1 + g [2 G_sigma(r) - 1]), where
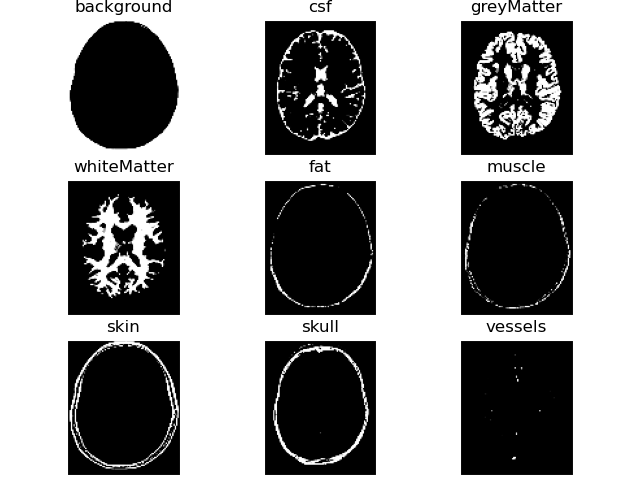
# show region probability masks
PetClass = brainweb.FDG
label_probs = brainweb.get_label_probabilities(files[-1], labels=PetClass.all_labels)
volshow(label_probs[brainweb.trim_zeros_ROI(label_probs)], titles=PetClass.all_labels, frameon=False);brainweb.seed(1337)
for f in tqdm(files, desc="mMR ground truths", unit="subject"):
vol = brainweb.get_mmr_fromfile(
f,
petNoise=1, t1Noise=0.75, t2Noise=0.75,
petSigma=1, t1Sigma=1, t2Sigma=1,
PetClass=PetClass)# show last subject
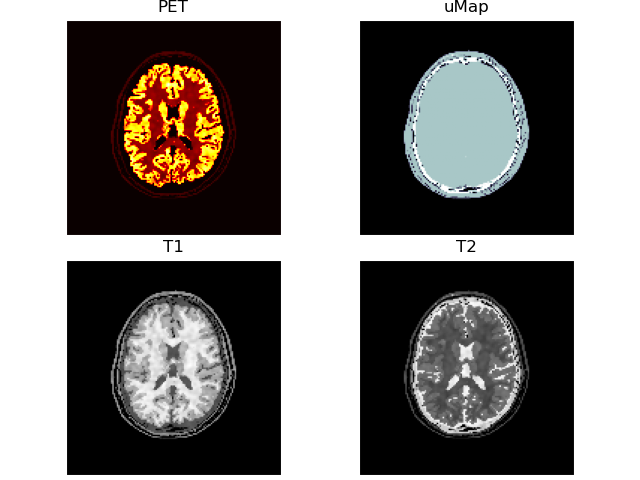
print(f)
volshow([vol['PET' ][:, 100:-100, 100:-100],
vol['uMap'][:, 100:-100, 100:-100],
vol['T1' ][:, 100:-100, 100:-100],
vol['T2' ][:, 100:-100, 100:-100]],
cmaps=['hot', 'bone', 'Greys_r', 'Greys_r'],
titles=["PET", "uMap", "T1", "T2"],
frameon=False);~/.brainweb/subject_54.bin.gz
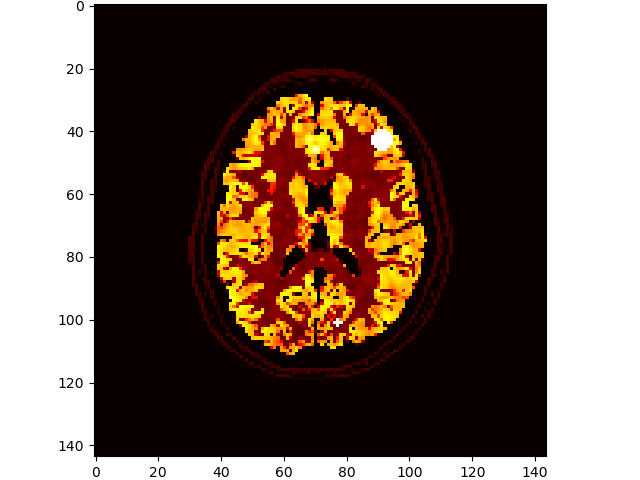
# add some lesions
brainweb.seed(1337)
im3d = brainweb.add_lesions(vol['PET'])
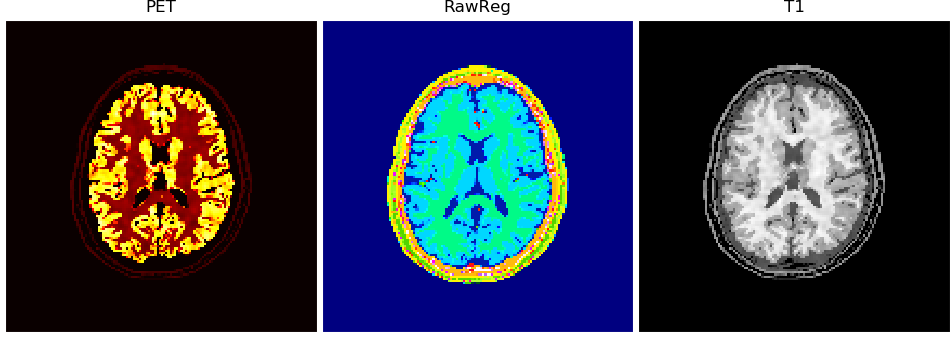
volshow(im3d[:, 100:-100, 100:-100], cmaps=['hot']);# bonus: use brute-force registration to transform
#!pip install -U 'brainweb[register]'
reg = brainweb.register(
data[:, ::-1], target=vol['PET'],
src_resolution=brainweb.Res.brainweb,
target_resolution=brainweb.Res.mMR)
volshow({
"PET": vol['PET'][:, 100:-100, 100:-100],
"RawReg": reg[ :, 100:-100, 100:-100],
"T1": vol['T1' ][:, 100:-100, 100:-100],
}, cmaps=['hot', 'gist_ncar', 'Greys_r'], ncols=3, tight_layout=5, figsize=(9.5, 3.5), frameon=False);