This repository contains all the analysis code to reproduce/explore the (currently) bioarxiv manuscript "The Latent Semantic Space and Corresponding Brain Regions of the Functional Neuroimaging Literature" doi: 10.1101/157826.
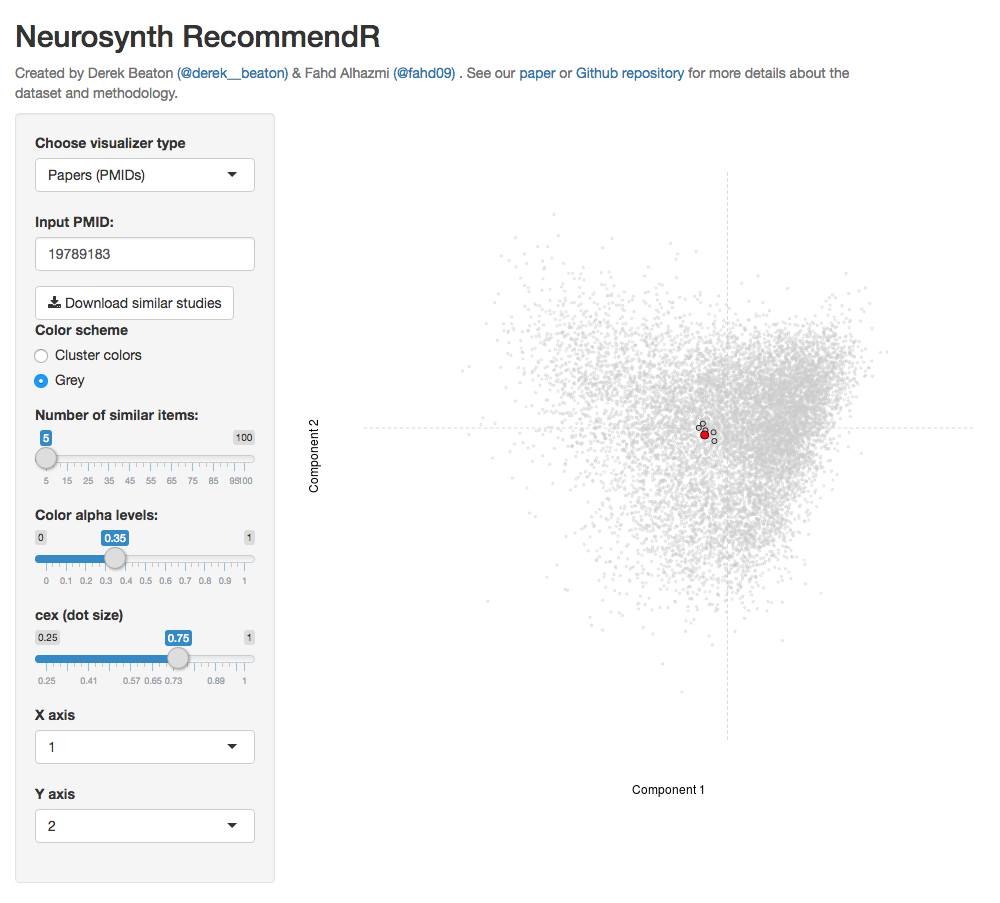
We recommend that you start with the Neurosynth RecommendR application that works as a recommendation engine for functional neuroimaging papers. This recommendation tool uses a distance-based search to retrieve papers (PMIDs) that are the most semantically similar to a given paper (PMID). You only need to provide: (1) a PMID of a paper of interest. Additionally, in the same way as the papers, you can also enter a term of interest and retrieve the closest terms.
In order to run the code and reproduce the work from A to Z, you need to download packages and libraries as required. The last successful run was conducted using R version 3.3.3 and Python 3.6.0 (through Anaconda enviroment).
Beside that, you will need to download this neurosynth dataset (5 February, 2015) and unzip the file, then move 'database.txt' to 'data/' folder here. This file is > 100MB and cannot be uploaded to Github. In cases you want to work with the latest Neurosynth data release, the script should work fine but you might see different results.
Hopefully, file names in 'script' folder are clear enough. You should run them in the same order if you want to run this project from scratch. However, you don't need to as most inputs/outputs are already in the data/ folder. Oh.. the document-term matrix (dtm) is 270MB and cannot be uploaded in this repo but you can easily generate it by running 2_preprocess_textmine.R, or, download it directly from here. Again, you should download it to the 'data' folder.
Just open a new issue and I will do my best to help.