-
Notifications
You must be signed in to change notification settings - Fork 0
EVE Project Data
EFV sequences were recovered from whole genome sequence (WGS) assemblies via database-integrated genome screening (DIGS) using the DIGS tool.
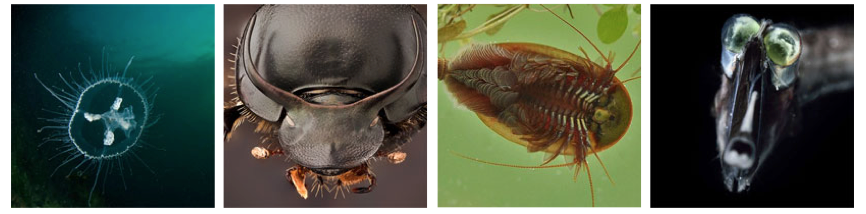
Species With EFVs Some of the species in which we identified novel endogenous flaviviral elements (EFVs) Left to right: freshwater jellyfish (Craspedacusta sowerbyi), long-horned beetle (Anoplophora glabripennis), tadpole shrimp (Lepidurus arcticus), tube-eye fish (Stylephorus chordatus).
Raw FASTA for EFVs recovered via DIGS are here.
Sequence-associated data in tabular format are here.
The tabular files contain information about the genomic locations of EFVs. All data pertaining to this screen are included in this repository.
- The complete list of vertebrate genomes screened can be found here.
- The complete list of invertebrate genomes screened can be found here.
- The set of flavivirus polypeptide sequences used as probes can be found here.
- The final set of flavivirus and EFV polypeptide sequences used as references can be found here.
- Input parameters for screening using the DIGS tool can be found here.
We constructed reference sequences for EFVs using alignments of EFV sequences derived from the same initial germline colonisation event – i.e., orthologous elements in distinct species, and paralogous elements that have arisen via intragenomic duplication of EFV sequences.
EFV consensus/reference FASTA is here.
Tabular formatted metadata for EFV reference sequences is here.
We have applied a systematic approach to naming endogenous flavivirids (EFVs), following a convention developed for endogenous retroviruses. Each individual EFV locus was assigned a unique identifier (ID) constructed from several components, each of which refers to a property of the locus.

The first component is the classifier ‘EFV’ (endogenous flavivirid).
The second component is a composite of two distinct subcomponents separated by a period:
(i) the name of the EFV group;
(ii) a numeric ID that uniquely identifies the insertion. The numeric ID is an integer that identifies a unique insertion locus that arose as a consequence of an initial germline infection. Thus, orthologous copies in different species are given the same number.
The third component of the ID defines the set of host species in which the ortholog occurs.