-
Notifications
You must be signed in to change notification settings - Fork 0
Core Project Data
The HCV-GLUE core project provides a minimal set of essential data components for performing comparative genomic analysis of parvoviruses.
The data items included in the core project are:
-
A set of HCV genome feature definitions covering all genera.
-
An annotated reference genome sequence for each HCV genotype and subtype (i.e. one species per genus).
-
A hierarchically arranged set of multiple sequence alignments representing sequence homology among HCV reference sequences.
-
A reference phylogeny
HCV has a monopartite genome ~10 kilobases (Kb) in length that encodes a large polyprotein that are co- and post-translationally cleaved to generate mature virus proteins. The structural proteins of the virion - capsid (C), premembrane (prM) and envelope (E) - are encoded toward the 5’ end of the genome, while genes encoding non-structural (NS) proteins are located further downstream.
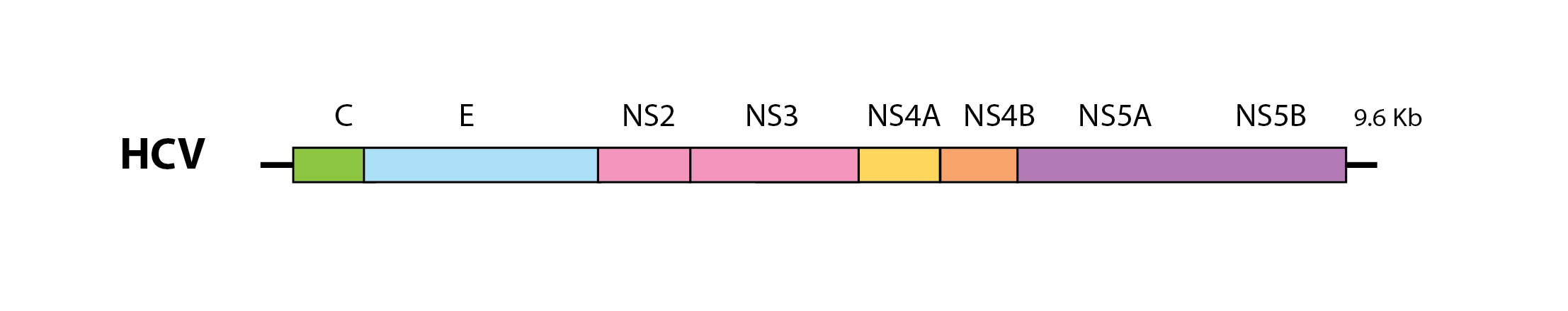
A schematic representation of the hepatitis C virus (HCV) genome. Kb=kilobases
In HCV-GLUE a standard set of HCV genome features is defined in this project build file.
HCV-GLUE contains reference sequences for HCV subtypes and genotypes.
Sequences are in GenBank XML format and are contained in this directory.
Reference sequences are defined in this csv file.
Clade definitions follow rules defined here.
A reference phylogeny was reconstructed to represent the evolutionary relationships between HCV reference sequences. The phylogeny was reconstructed using RAXML, via GLUE's raxmlPhylogenyGenerator module.
Phylogenies are here, a pdf version of the tree can be obtained/viewed here.