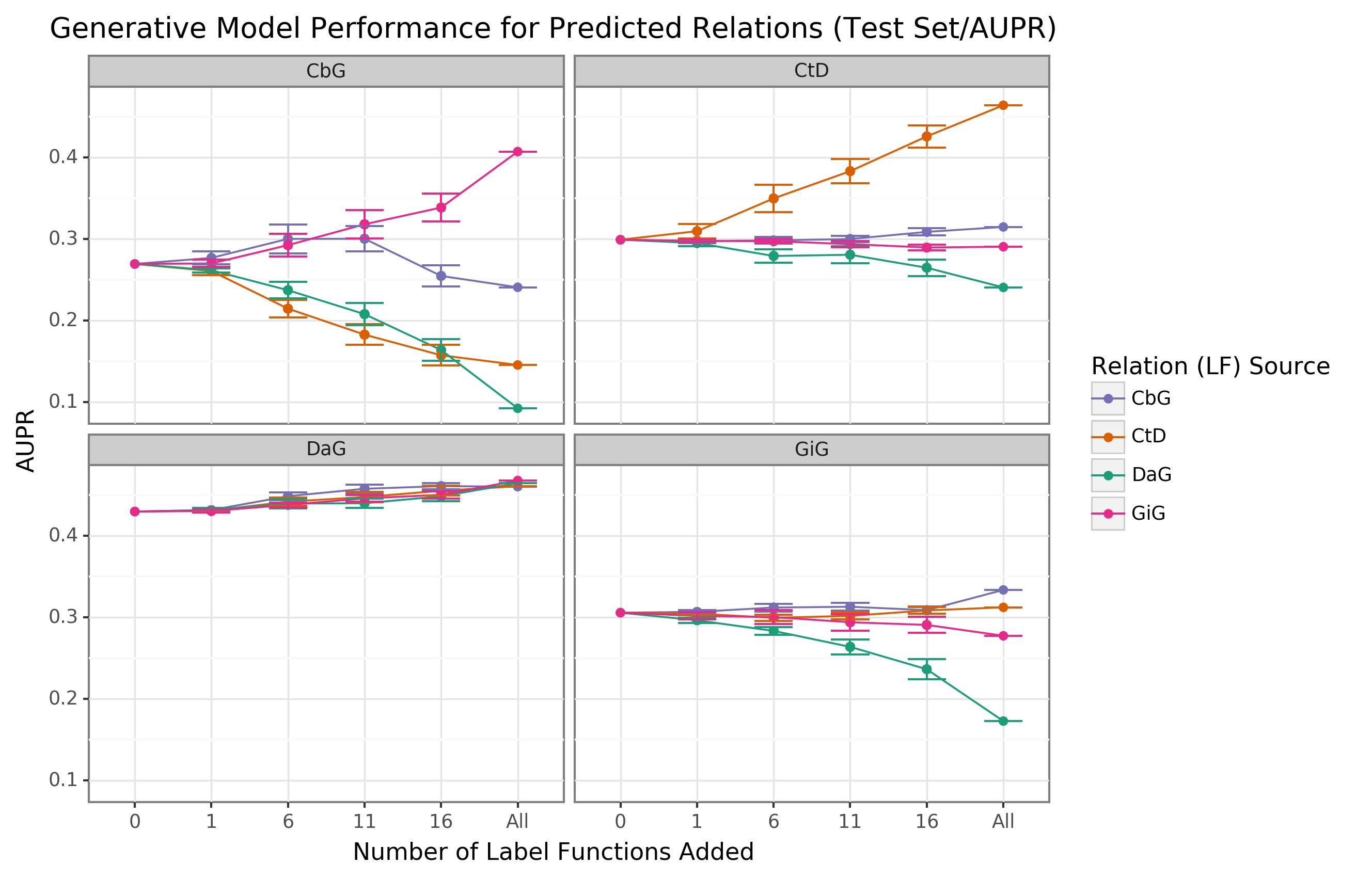
 {#fig:aupr_gen_model_test_set}
{#fig:aupr_gen_model_test_set}
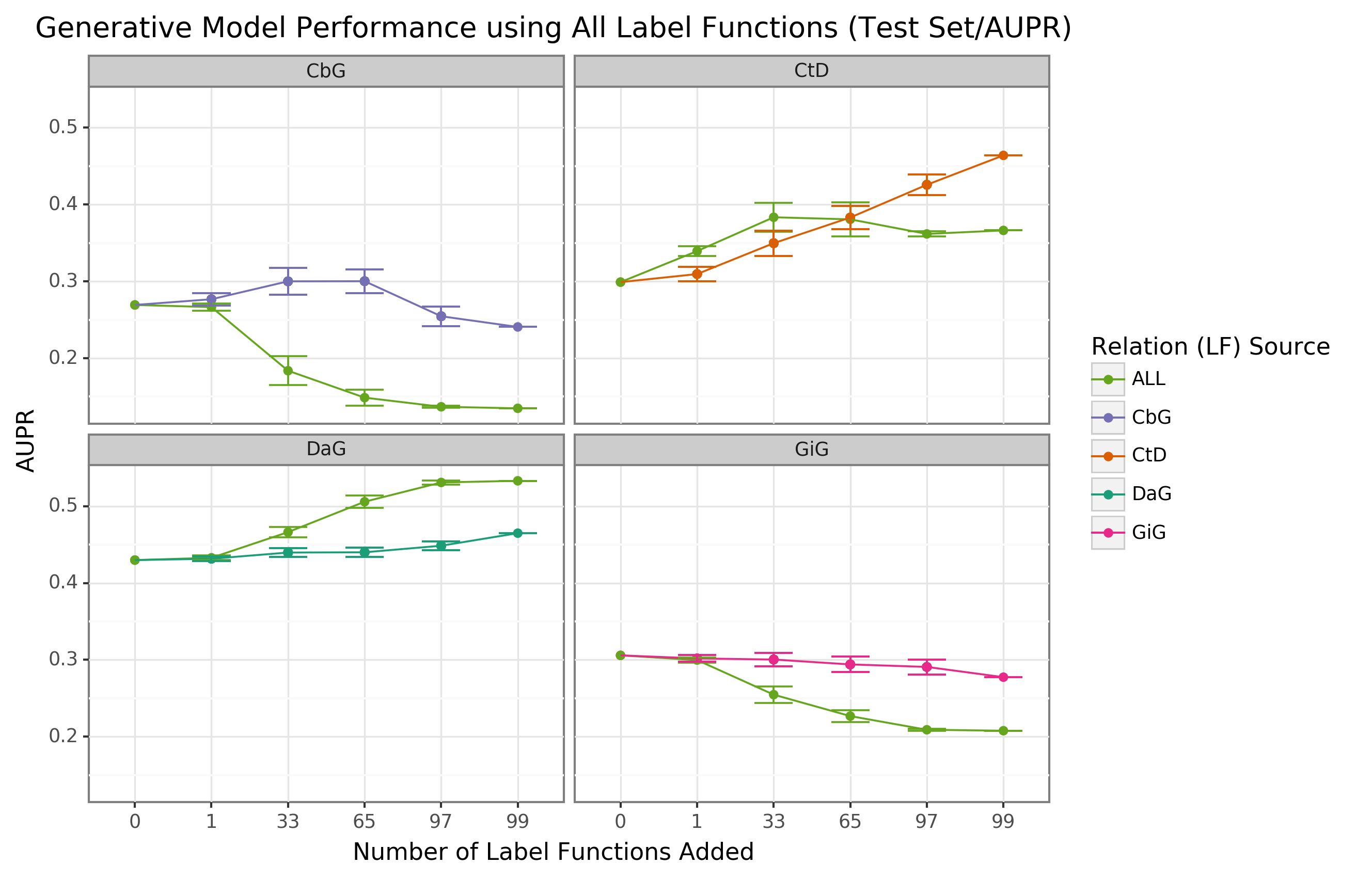 {#fig:aupr_grabbag_gen_model_test_set}
{#fig:aupr_grabbag_gen_model_test_set}
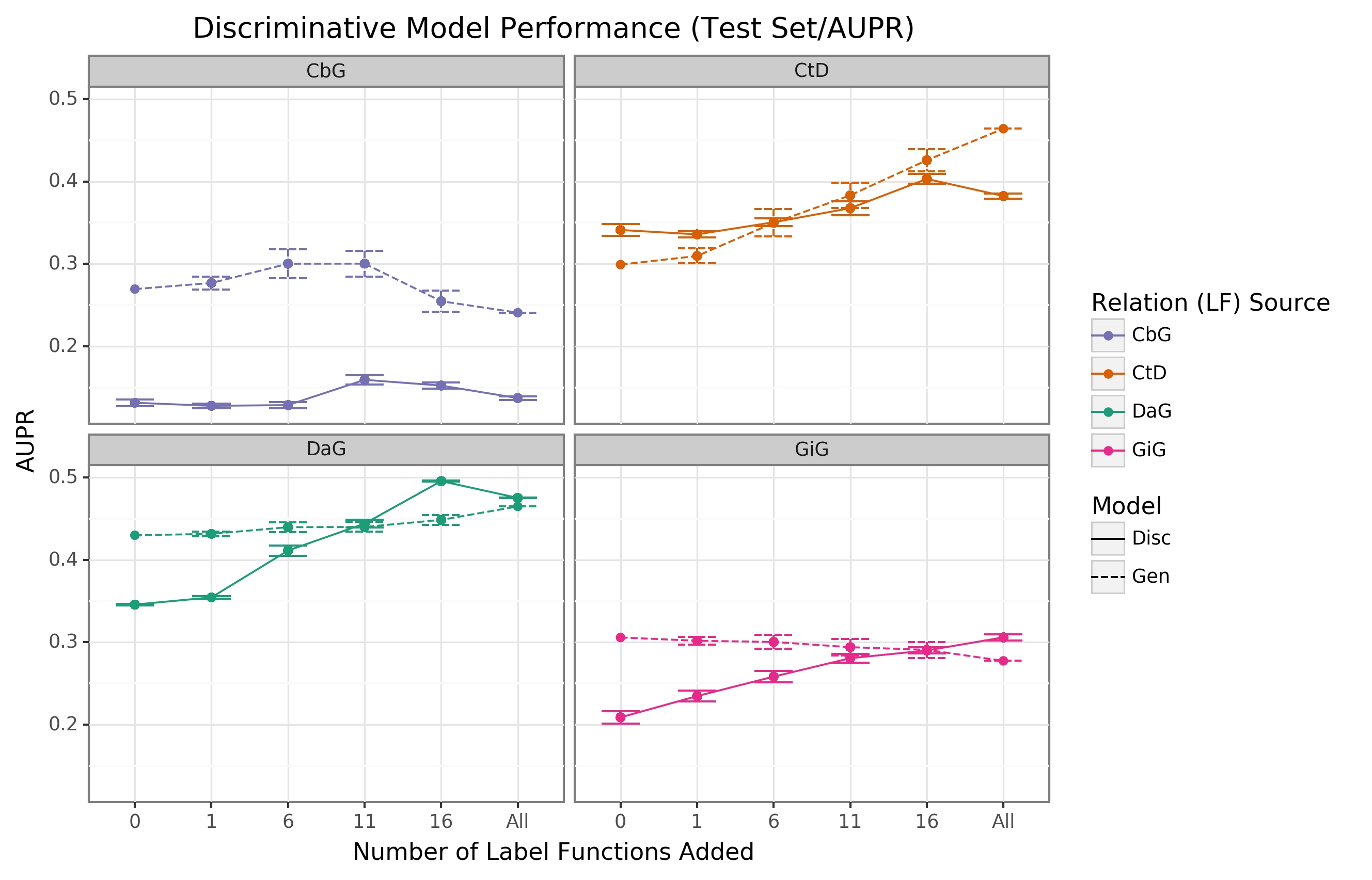 {#fig:aupr_discriminative_model_performance}
{#fig:aupr_discriminative_model_performance}
| Edge Type | Source Node | Target Node | Generative Model Prediction | Discriminative Model Prediction | Number of Sentences | In Hetionet | Text |
|---|---|---|---|---|---|---|---|
| [D]{.disease_color}a[G]{.gene_color} | hematologic cancer | STMN1 | 1.000 | 0.979 | 83 | Novel | the stathmin1 mrna expression level in de novo al patient be high than that in healthy person ( p < 0.05 ) , the [stathmin1].{gene_color} mrna expression level in relapse patient with al be high than that in de novo patient ( p < 0.05 ) , and there be no significant difference of stathmin1 mrna expression between patient with [aml].{disease_color} and patient with all . |
| [D]{.disease_color}a[G]{.gene_color} | breast cancer | INSIG2 | 1.000 | 0.979 | 4 | Novel | in analysis of [idc ].{disease_color} cell , the level of [insig2].{gene_color} mrna expression be significantly high in late - stage patient than in early - stage patient . |
| [D]{.disease_color}a[G]{.gene_color} | lung cancer | GNAO1 | 1.000 | 0.979 | 104 | Novel | high [numb].{disease_color} expression be associate with favorable prognosis in patient with [lung adenocarcinoma].{gene_color} , but not in those with squamous cell carcinoma . |
| [D]{.disease_color}a[G]{.gene_color} | breast cancer | TTF1 | 1.000 | 0.977 | 88 | Novel | significant [ttf-1].{gene_color} overexpression be observe in adenocarcinomas harbor egfr mutation ( p = 0.008 ) , and no or significantly low level expression of ttf-1 be observe in [adenocarcinomas].{disease_color} harbor kras mutation ( p = 0.000 ) . |
| [D]{.disease_color}a[G]{.gene_color} | breast cancer | BUB1B | 1.000 | 0.977 | 13 | Novel | elevated [bubr1].{gene_color} expression be associate with poor survival in early stage [breast cancer].{disease_color} patient . |
| [D]{.disease_color}a[G]{.gene_color} | Alzheimer's disease | SERPINA3 | 1.000 | 0.977 | 182 | Existing | a common polymorphism within act and il-1beta gene affect plasma level of [act].{gene_color} or il-1beta , and [ad].{disease_color} patient with the act t , t or il-1beta t , t genotype show the high level of plasma act or il-1beta , respectively . |
| [D]{.disease_color}a[G]{.gene_color} | esophageal cancer | TRAF6 | 1.000 | 0.976 | 15 | Novel | expression of traf6 be highly elevated in [esophageal cancer].{disease_color} tissue , and patient with high [traf6].{gene_color} expression have a significantly short survival time than those with low traf6 expression . |
| [D]{.disease_color}a[G]{.gene_color} | hypertension | TBX4 | 1.000 | 0.975 | 146 | Novel | the proportion of circulate [th1].{gene_color} cell and the level of t - bet , ifng mrna be increase in [ht].{disease_color} patient , the expression of ifng - as1 be upregulated and positively correlate with the proportion of circulate th1 cell or t - bet , and ifng expression , or serum level of anti - thyroglobulin antibody / thyroperoxidase antibody in ht patient . |
| [D]{.disease_color}a[G]{.gene_color} | breast cancer | TP53 | 1.000 | 0.975 | 3481 | Existing | hormone receptor status rather than her2 status be significantly associate with increase ki-67 and [p53].{gene_color} expression in triple [- negative ].{disease_color} breast carcinoma , and high expression of ki-67 but not p53 be significantly associate with axillary nodal metastasis in triple - negative and high - grade non - triple - negative breast carcinoma . |
| [D]{.disease_color}a[G]{.gene_color} | esophageal cancer | COL17A1 | 1.000 | 0.975 | 32 | Novel | high [cd147].{gene_color} expression in patient with [esophageal cancer].{disease_color} be associate with bad survival outcome and common clinicopathological indicator of poor prognosis . |
| [C]{.compound_color}t[D]{.disease_color} | Docetaxel | prostate cancer | 0.996 | 0.964 | 5614 | Existing | docetaxel and atrasentan versus [docetaxel ].{compound_color} and placebo for man with advanced castration - resistant [prostate cancer].{disease_color} ( swog s0421 ) : a randomised phase 3 trial |
| [C]{.compound_color}t[D]{.disease_color} | E7389 | breast cancer | 0.999 | 0.957 | 862 | Novel | clinical effect of prior trastuzumab on combination [eribulin mesylate].{compound_color} plus trastuzumab as first - line treatment for human epidermal growth factor receptor 2 positive locally recurrent or metastatic [breast cancer].{disease_color} : result from a phase ii , single - arm , multicenter study |
| [C]{.compound_color}t[D]{.disease_color} | Zoledronate | bone cancer | 0.996 | 0.955 | 226 | Novel | [zoledronate].{compound_color} in combination with chemotherapy and surgery to treat [osteosarcoma].{disease_color} ( os2006 ) : a randomised , multicentre , open - label , phase 3 trial . |
| [C]{.compound_color}t[D]{.disease_color} | 0.878 | 0.954 | 484 | Existing | the role of [ixazomib].{compound_color} as an augment conditioning therapy in salvage autologous stem cell transplant ( asct ) and as a post - asct consolidation and maintenance strategy in patient with relapse multiple myeloma ( accord [ uk - mra [myeloma].{disease_color} xii ] trial ) : study protocol for a phase iii randomise controlled trial | ||
| [C]{.compound_color}t[D]{.disease_color} | Topotecan | lung cancer | 1.000 | 0.954 | 315 | Existing | combine chemotherapy with cisplatin , etoposide , and irinotecan versus [topotecan].{compound_color} alone as second - line treatment for patient with [sensitive relapse small].{disease_color} - cell lung cancer ( jcog0605 ) : a multicentre , open - label , randomised phase 3 trial . |
| [C]{.compound_color}t[D]{.disease_color} | Epirubicin | breast cancer | 0.999 | 0.953 | 2147 | Existing | accelerate versus standard [epirubicin].{compound_color} follow by cyclophosphamide , methotrexate , and fluorouracil or capecitabine as adjuvant therapy for [breast cancer].{disease_color} in the randomised uk tact2 trial ( cruk/05/19 ) : a multicentre , phase 3 , open - label , randomise , control trial |
| [C]{.compound_color}t[D]{.disease_color} | Paclitaxel | breast cancer | 1.000 | 0.952 | 10255 | Existing | sunitinib plus [paclitaxel].{compound_color} versus bevacizumab plus paclitaxel for first - line treatment of patients with [advanced breast cancer].{disease_color} : a phase iii , randomized , open - label trial |
| [C]{.compound_color}t[D]{.disease_color} | Anastrozole | breast cancer | 0.996 | 0.952 | 2364 | Existing | a european organisation for research and treatment of cancer randomize , double - blind , placebo - control , multicentre [phase].{disease_color} ii trial of anastrozole in combination with [gefitinib or placebo in hormone].{compound_color} receptor - positive advanced breast cancer ( nct00066378 ) . |
| [C]{.compound_color}t[D]{.disease_color} | Gefitinib | lung cancer | 1.000 | 0.950 | 11860 | Existing | [gefitinib].{compound_color} versus placebo as maintenance therapy in patient with locally advanced or metastatic [non - small].{disease_color} - cell lung cancer ( inform ; c - tong 0804 ) : a multicentre , double - blind randomise phase 3 trial . |
| [C]{.compound_color}t[D]{.disease_color} | Docetaxel | prostate cancer | 1.000 | 0.949 | 5614 | Existing | ipilimumab versus placebo after radiotherapy in patient with metastatic castration - resistant [prostate cancer].{disease_color} that have progress after [docetaxel].{compound_color} chemotherapy ( ca184 - 043 ) : a multicentre , randomised , double - blind , phase 3 trial |
| [C]{.compound_color}t[D]{.disease_color} | Sulfamethazine | lung cancer | 0.611 | 0.949 | 4 | Novel | [tmp].{compound_color} / smz ( 320/1600 mg / day ) treatment be compare to placebo in a double - blind , randomized trial in [patient with newly diagnose].{disease_color} small cell carcinoma of the lung during the initial course of chemotherapy with cyclophosphamide , doxorubicin , and etoposide . |
| [C]{.compound_color}b[G]{.gene_color} | D-Tyrosine | EGFR | 0.601 | 0.876 | 3423 | Novel | amphiregulin ( ar ) and heparin - binding egf - like growth factor ( hb - [egf].{gene_color} ) bind and activate the egfr while heregulin ( hrg [) act ].{compound_color} through the p185erbb-2 and p180erbb-4 tyrosine kinase . |
| [C]{.compound_color}b[G]{.gene_color} | Phosphonotyrosine | ANK3 | 0.004 | 0.865 | 1 | Novel | at least two domain of p85 can bind to [ank3 ].{gene_color} , and the interaction involve the p85 c - sh2 domain be find to be [phosphotyrosine].{compound_color} - independent . |
| [C]{.compound_color}b[G]{.gene_color} | Adenosine | ABCC8 | 0.891 | 0.860 | 353 | Novel | sulfonylurea act by inhibition of [beta - cell ].{compound_color} adenosine triphosphate - dependent potassium ( k(atp ) ) channel after bind to the sulfonylurea subunit 1 [receptor ( ].{gene_color} sur1 ) . |
| [C]{.compound_color}b[G]{.gene_color} | D-Tyrosine | AREG | 0.891 | 0.857 | 22 | Novel | amphiregulin ( [ar ) ].{gene_color} and heparin - binding egf - like growth factor ( hb - egf ) bind and activate the egfr while heregulin ( hrg [) act ].{compound_color} through the p185erbb-2 and p180erbb-4 tyrosine kinase . |
| [C]{.compound_color}b[G]{.gene_color} | D-Tyrosine | EGF | 0.602 | 0.856 | 389 | Novel | upon activation of the receptor for the epidermal growth factor ( [egfr ) ].{gene_color} , sprouty2 undergoe phosphorylation at a conserve [tyrosine ].{compound_color} that recruit the src homology 2 domain of c - cbl . |
| [C]{.compound_color}b[G]{.gene_color} | D-Tyrosine | CSF1 | 0.101 | 0.854 | 106 | Novel | as a member of the subclass iii family of receptor [tyrosine].{compound_color} kinase , kit be closely relate to the receptor for platelet derive growth factor alpha and beta ( pdgf - a and b [) , macrophage colony ].{gene_color} stimulate factor ( m - csf ) , and flt3 ligand . |
| [C]{.compound_color}b[G]{.gene_color} | D-Tyrosine | ERBB4 | 0.101 | 0.848 | 115 | Novel | the efgr family be a group of four structurally similar [tyrosine ].{compound_color} kinase ( egfr , her2 / neu , erbb-3 [, and erbb-4].{gene_color} ) that dimerize on bind with a number of ligand , include egf and transform growth factor alpha . |
| [C]{.compound_color}b[G]{.gene_color} | D-Tyrosine | EGFR | 0.969 | 0.848 | 3423 | Novel | the [epidermal growth factor receptor ].{gene_color} be a member of type - -pron- growth factor receptor [family ].{compound_color} with tyrosine kinase activity that be activate follow the binding of multiple cognate ligand . |
| [C]{.compound_color}b[G]{.gene_color} | D-Tyrosine | VAV1 | 0.601 | 0.842 | 187 | Novel | stimulation of quiescent rodent fibroblast with either epidermal or platelet - derive growth factor induce an increase affinity of vav for cbl - b and result in the [subsequent ].{gene_color} formation of a vav - [dependent ].{compound_color} trimeric complex with the ligand - stimulate tyrosine kinase receptor . |
| [C]{.compound_color}b[G]{.gene_color} | Tretinoin | RORB | 0.601 | 0.840 | 7 | Novel | the retinoid z receptor beta ( [rzr beta ) ].{gene_color} , an orphan receptor , be a member of the [retinoic acid].{compound_color} receptor ( rar)/thyroid hormone receptor ( tr ) subfamily of nuclear receptor . |
| [C]{.compound_color}b[G]{.gene_color} | L-Tryptophan | TACR1 | 0.891 | 0.839 | 4 | Novel | these result suggest that the [tryptophan ].{compound_color} and quinuclidine series of nk-1 antagonist bind to similar bind site on the human [nk-1 receptor ].{gene_color} . |
| [G]{.gene_color}i[G]{.gene_color} | CYSLTR2 | CYSLTR2 | 0.967 | 0.564 | 37 | Novel | the bind pocket of [cyslt2 ].{gene2_color} receptor and the proposition of the interaction mode between [cyslt2 ].{gene1_color} and hami3379 be identify . |
| [G]{.gene_color}i[G]{.gene_color} | RXRA | PPARA | 1.000 | 0.563 | 143 | Novel | after bind ligand , the [ppar ].{gene2_color} - y receptor heterodimerize [with ].{gene1_color} the rxr receptor . |
| [G]{.gene_color}i[G]{.gene_color} | RXRA | RXRA | 0.824 | 0.551 | 1101 | Existing | nuclear hormone receptor , for example , bind either as homodimer or as heterodimer with [retinoid x receptor ].{gene1_color} ( [rxr ) ].{gene2_color} to half - site repeat that be stabilize by protein - protein interaction mediate by residue within both the dna- and ligand - bind domain . |
| [G]{.gene_color}i[G]{.gene_color} | ADRBK1 | ADRA2A | 0.822 | 0.543 | 3 | Novel | mutation of these residue within the [holo - alpha(2a)ar diminish grk2-promoted].{gene2_color} phosphorylation [of ].{gene1_color} the receptor as well as the ability of the kinase to be activate by receptor binding . |
| [G]{.gene_color}i[G]{.gene_color} | ESRRA | ESRRA | 0.001 | 0.531 | 308 | Existing | the crystal structure of the ligand bind domain ( lbd ) of the estrogen - relate receptor [alpha ].{gene2_color} ( [erralpha , ].{gene1_color} nr3b1 ) complexe with a coactivator peptide from peroxisome proliferator - activate receptor coactivator-1alpha ( pgc-1alpha ) reveal a transcriptionally active conformation in the absence of a ligand . |
| [G]{.gene_color}i[G]{.gene_color} | GP1BA | VWF | 0.518 | 0.527 | 144 | Existing | these finding indicate the novel bind site require for [vwf ].{gene2_color} binding of human [gpibalpha ].{gene1_color} . |
| [G]{.gene_color}i[G]{.gene_color} | NR2C1 | NR2C1 | 0.027 | 0.522 | 26 | Novel | the human [testicular receptor 2].{gene1_color} ( [tr2 )].{gene2_color} , a member of the nuclear hormone receptor superfamily , have no identify ligand yet . |
| [G]{.gene_color}i[G]{.gene_color} | NCOA1 | ESRRG | 0.992 | 0.518 | 1 | Novel | the crystal structure of the ligand bind domain ( lbd ) of the estrogen - relate receptor [3 (].{gene2_color} err3 ) complexe with a steroid receptor [coactivator-1 (].{gene1_color} src-1 ) peptide reveal a transcriptionally active conformation in absence of any ligand . |
| [G]{.gene_color}i[G]{.gene_color} | PPARG | PPARG | 0.824 | 0.504 | 2497 | Existing | although these agent can bind and activate an orphan nuclear receptor , [peroxisome proliferator - activate].{gene2_color} receptor [gamma ( ].{gene1_color} ppargamma ) , there be no direct evidence to conclusively implicate this receptor in the regulation of mammalian glucose homeostasis . |
| [G]{.gene_color}i[G]{.gene_color} | ESR2 | ESR1 | 0.995 | 0.503 | 1715 | Novel | ligand bind experiment with purify [er alpha].{gene2_color} and [er beta].{gene1_color} confirm that the two phytoestrogen be er ligand . |
| [G]{.gene_color}i[G]{.gene_color} | FGFR2 | FGFR2 | 1.000 | 0.501 | 584 | Existing | receptor modeling of [kgfr].{gene1_color} be use to identify selective kgfr tyrosine kinase ( tk ) inhibitor molecule that have the potential to bind selectively to the [kgfr].{gene2_color} . |
| Table: Contains the top ten predictions for each edge type. Highlighted words represent entities mentioned within the given sentence. {#tbl:edge_prediction_tbl} |