Python 3 Dependencies: csv, argparse, cairosvg This program is a little memory hungry... Note: Current version uses the GRCh37 (hg19) genome. If a different species or genome build is desired, the "chromosome_sizes" list can be adjusted in the python skript. A bar with ploidies will be plotted on the right side of the heatmap. Usage: -o Output filename -c node_unmerged_cnv_calls.bed file from the 10x pipeline output -p per_cell_summary_metrics.csv file from the 10x pipeline output Optional: -e Set to "no" to include noisy cells marked by the 10x pipeline. Default is "yes" -f Set a lower ploidy cuttoff if desired. E.g "-f 1.1" will exclude all cells with a ploidy lower than 1.1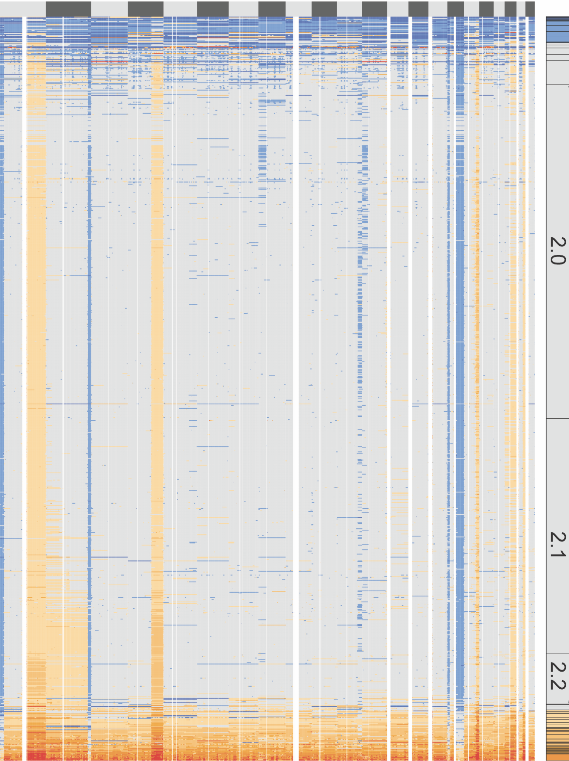
forked from StefanKurtenbach/scCNV_heatmap
-
Notifications
You must be signed in to change notification settings - Fork 0
harbourlab/scCNV_heatmap
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
About
Generates meaninful heatmaps from 10x single cell CNV data
Resources
Stars
Watchers
Forks
Packages 0
No packages published
Languages
- Python 100.0%