pysagem is a CLI application that lets you simulate biological invasions in dynamic landscapes. By providing an animation flag, it also lets you visualise the simulation. The name pysagem is a direct borrowing from the Portuguese paisagem, which means landscape and has a somewhat similar pronunciation.
- numpy
- scipy
- numba
- matplotlib
- palettable
- alive_progress<2.2
Also, for the animation to be generated, you need to have ffmpeg installed.
pysagem can be installed with pip:
pip install pysagem
usage: pysagem [--parameter <value>] [options]
_ __ _ _ ___ __ _ __ _ ___ _ __ ___
| '_ \| | | / __|/ _` |/ _` |/ _ \ '_ ` _ \
| |_) | |_| \__ \ (_| | (_| | __/ | | | | |
| .__/ \__, |___/\__,_|\__, |\___|_| |_| |_|
| | __/ | __/ |
|_| |___/ |___/
Simulate and visualize biological invasions in dynamic landscapes
Arguments:
Initial Disturbance:
--p_initialdist <float> disturbance intensity, i.e., the probability of a random
patch being selected to be disturbed (default: 0.5)
--q_initialdist <float> disturbance degree of clustering (default: 1.0)
Restorations:
--rest enables restorations (default: False)
--p_rest <float> restoration intensity, i.e., the probability of a random
patch being selected to be restored (default: 0.5)
--n_rest <int> number of restorations (default: 5)
Additional Disturbances:
--add_dist enables additional disturbances (default: False)
--p_add_dist <float> additional disturbance intensity (default: 0.5)
--q_add_dist <float> degree of clustering of additional disturbance (default: 1.0)
--n_add_dist <int> number of additional disturbances (default: 5)
General:
--length <int> length of the square matrix (default: 50)
--gen <int> number of generations (default: 1000)
--radius <float> radius of neighbors to consider (default: 3.0)
--alpha <float> effect of the invasive species on the native species (default: 0.8)
--beta <float> effect of the native species on the invasive species (default: 0.8)
--rn <float> growth rate of the native species (default: 1.0)
--ri <float> growth rate of the invasive species (default: 1.0)
--invasors <int> number of invasors (default: 1000)
--mignative <float> migration rate of native species (default: 0.2)
--miginvasive <float> migration rate of invasive species (default: 0.2)
Other Options:
-h, --help show this help message and exit
-s <int>, --seed <int> seed for reproducibility (default: random int)
-a, --animation generates an animation according to the simulation
-n <str>, --name <str> name of the run used for outputs (default: pysagem)
To run a simulation with the default arguments just type pysagem in the command line.
Arguments can either be specified directly on the command line or by providing a parameter file. A parameter file can be supplied using @ on the command line. The file must follow the structure of one space-delimited argument per line:
--argumentX valueX
--argumentY valueY
--argumentZ
Take this parameter file named params.txt as an example:
--seed 123
--gen 100
--p_initialdist 0.1
--q_initialdist 0.9
--rest
--n_rest 4
--add_dist
--n_add_dist 2
--mignative 0.9
--miginvasive 0.8
--animation
Running the command pysagem @params.txt:
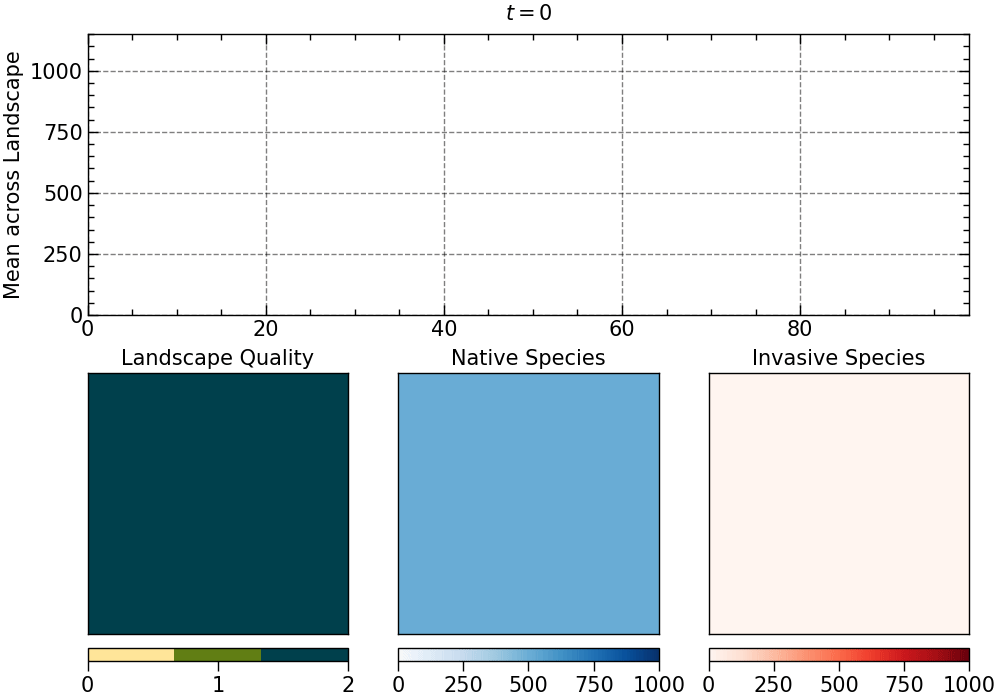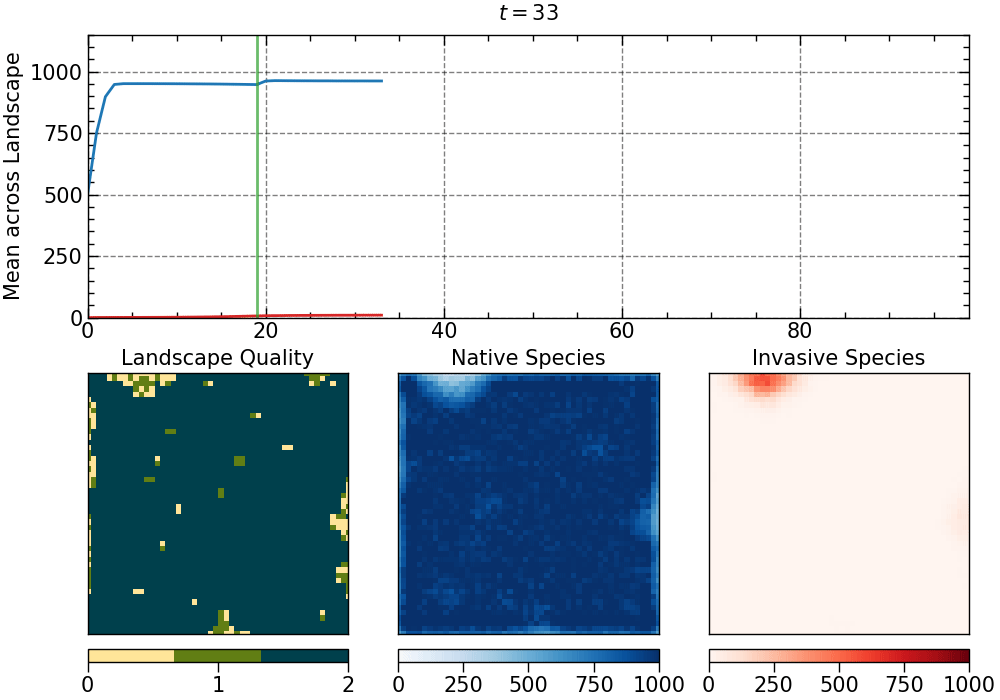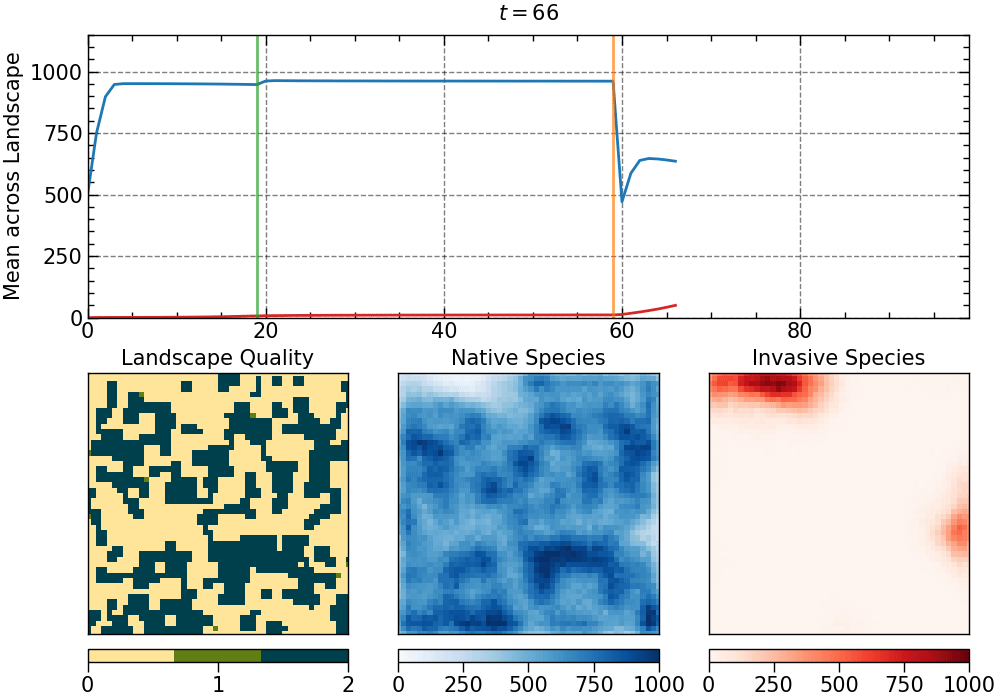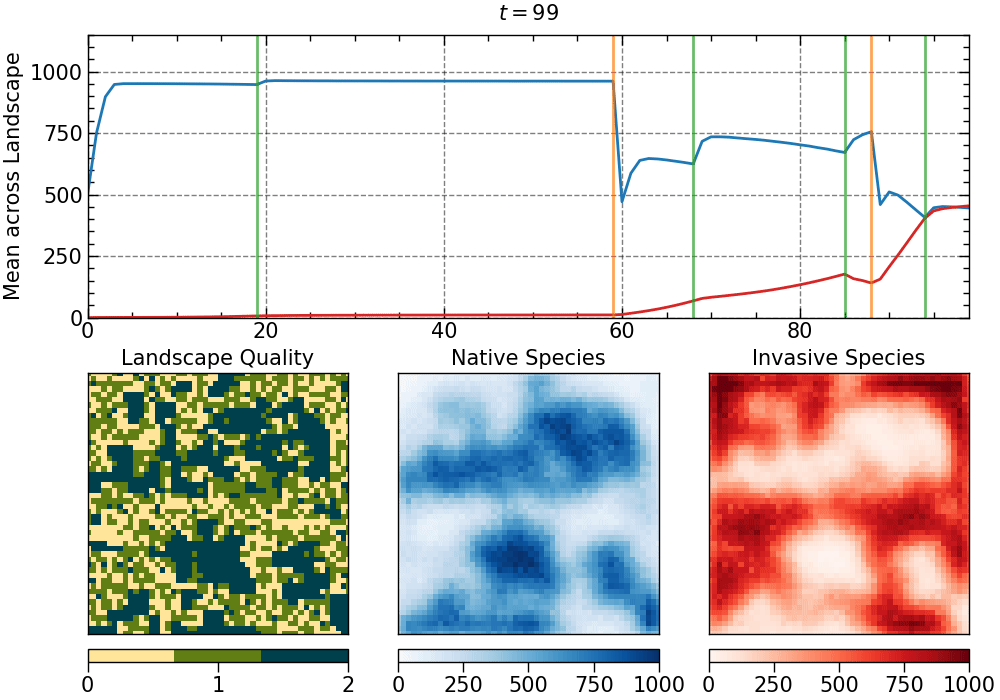
The animation produced for this choice of parameters:
The animation is saved as an mp4 file. The length of the animation will depend on the number of generations.
The simulation parameters are stored in a log file in the current working directory. The log file contains the parameters used for the simulation and the events that occurred during the simulation. The log file for the previous example would look like this:
# run name: pysagem
general parameters:
• seed: 123
• matrix size: 50x50
• number of generations: 100
• neighbors radius: 3.0
• number of invasors: 1000
• native species migration rate: 0.9
• invasive species migration rate: 0.8
• alpha: 0.8
• beta: 0.8
• rn: 1.0
• ri: 1.0
events:
• initial disturbance:
- intensity: 0.1
- degree of clustering: 0.9
- aggregate pattern
• restorations:
- intensity: 0.5
- random pattern
- selected generations:
[19, 68, 85, 94]
• additional disturbances:
- intensity: 0.5
- degree of clustering: 1.0
- aggregate pattern
- selected generations:
[59, 88]
The simulation generates a .npz file named after the run name. The file contains the following arrays:
natpop: the native species population matrix for each generation.invpop: the invasive species population matrix for each generation.landscape: the landscape matrix for each generation.mean_nat: the mean of the native species population for each generation.mean_inv: the mean of the invasive species population for each generation.
The output can be accessed using the numpy library:
import numpy as np
data = np.load('pysagem.npz')
natpop = data['natpop']
print(natpop.shape) # (number of generations, number of patches)
# to access the native species population matrix for the first generation
print(natpop[0])
print(natpop[0].shape) # (number of patches,)
# to acess the number of individuals of the native species in the patch 7 for the first generation
print(natpop[0][7]) # you can index using natpop[i][j] or natpop[i, j]
landscape = data['landscape']
print(landscape.shape) # (number of generations, number of patches)
# to acess the landscape matrix for the last generation
print(landscape[-1])
print(landscape[-1].shape) # (number of patches,)
# to acess the quality of the patch 15 for the fifth generation
print(landscape[4][15])
mean_nat = data['mean_nat']
print(mean_nat.shape) # (number of generations,)
# to acess the mean of the native species population for the 10th generation
print(mean_nat[9])Although the goal of this package is to provide a simple way to simulate and visualize biological invasions from the command line, it is possible to customize the simulation by modifying the source code. The pysagem package is organized in a way that makes it easy to understand and modify the simulation parameters and events. The pysagem package is organized as follows:
src/
└── pysagem/
├── __init__.py
├── main.py
├── events.py
├── animation.py
├── plotstyle.mplstyle
└── utils.py
- The
main.pyfile contains the main simulation loop. - The
events.pyfile contains the functions that define the events that can occur during the simulation. - The
animation.pyfile contains the functions that generate the animation. - The
cli.pyfile contains the functions that parse the command line arguments and the parameter file. - The
plotstyle.mplstylefile is a custom matplotlib style sheet for the animation plot. - The
utils.pyfile contains some helper functions for seeding the numba random number generator, writing the log file, and other utility functions.
This project is under the GNU General Public License v3 (GPLv3).