-
Notifications
You must be signed in to change notification settings - Fork 7
Home

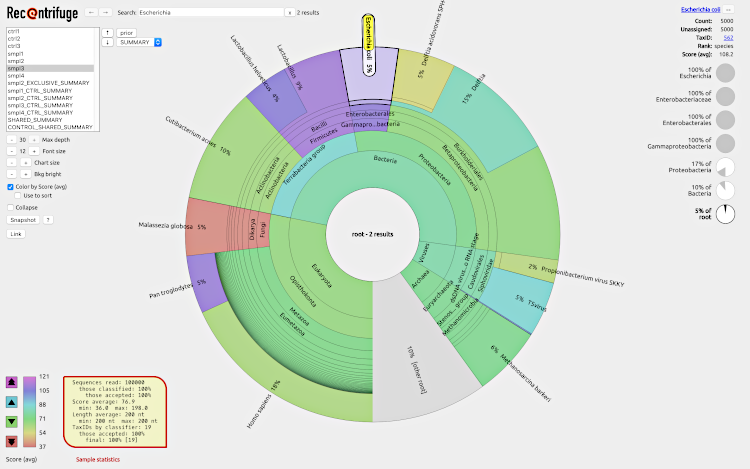
With Recentrifuge, researchers can analyze results from Centrifuge, LMAT, CLARK, Kraken, and other taxonomic classifiers using interactive charts with emphasis on the confidence level of the classifications. In addition to contamination-subtracted samples, Recentrifuge provides shared and exclusive taxa per sample, thus enabling robust contamination removal and comparative analysis in metagenomics.
Recentrifuge's novel approach combines robust statistics, arithmetic of scored taxonomic trees, and parallel computational algorithms. The software implements a robust method for the removal of negative-control and crossover taxa from the rest of samples.
Researchers use Recentrifuge to analyze very diverse metagenomic datasets. However, it is especially useful in the case of low microbial biomass metagenomic studies and when a more reliable detection of minority organisms is needed, like in clinical, environmental and forensic analysis.
- For installation of the software, please see the installation page.
- For instructions about how to run the main Recentrifuge script:
- if you have results from Centrifuge, please see running Recentrifuge for Centrifuge,
- in case you have LMAT outputs, your choice is running Recentrifuge for LMAT,
- if you have CLARK(S) results, please enter running Recentrifuge for CLARK,
- in case you have data from Kraken, please check running Recentrifuge for Kraken.
- if you use any other taxonomic classifier, please see running Recentrifuge for a generic classifier.
- For a comprehensive guide to the Recentrifuge options and flags, please check the Recentrifuge command line.
- For instructions about how to run the sequence extractor, please see running Rextract.
- For details about testing the different parts of the package, please enter testing.
- For an experimental feature related to infraspecific taxonomic ranks, please see the advanced flag --strain.
- For other advanced issues related to Recentrifuge you may consult extra topics in Troubleshooting.
This wiki also contains a page with some tips and tricks about Centrifuge:
- To generate your updated nt database, please check Centrifuge nt database.
To play with an example of a webpage generated by Recentrifuge, click on the screenshot:
If you use Recentrifuge in your research, please consider citing the paper. Thanks!
Martí JM (2019) Recentrifuge: Robust comparative analysis and contamination removal for metagenomics. PLOS Computational Biology 15(4): e1006967. https://doi.org/10.1371/journal.pcbi.1006967