We will use Biopython to handle biological sequence data stored in FASTA & PDB (Protein Data Bank) and XML format. Using this sequence data, we will explore and create an interactive three-dimensional (3D) representation of SARS-CoV-2 (Coronavirus) protein structures.
Data contains the genetic information in FASTA file format, downloaded from from NCBI (National Center for Biotechnology Information).
- Python 3.*
- Libraries: Biopython, Pylab, Pandas, urllib, nglview
- Jupyter Notebook
- Datasets: The complete set of files is publicly available and can be downloaded from the NCBI (National Center for Biotechnology Information).. Alternatively, you can find the folder (titled SARS-CoV-2-spike-protein-visualization) in my Github repository here.
- Others: The code can be run in JupyterLab as an Interactive Python Notebook (ipynb). No additional installation is required.
- JupyterLab allows you to use and share Jupyter notebooks with others without having to download, install, or run anything on your own computer (other than a browser).
In the fields of Bioinformatics, Health and Medical Technology & Biotechnology, there is now a widespread need for visualization tools to present the 3D structure of proteins. There are only a few examples of protein function. A remarkable fact is that all tasks they can perform are based on a common principle, the twenty amino acids that can form a protein. That is the reason why studying proteins, their composition, structure, dynamics and function, is so important.
We must understand how these molecules fold, how they assemble into complexes, how they function if we wish to answer questions such as why we have cancer, why we grow old, why we get sick, how can we find cures for many diseases, why life as we know it has evolved in this way and on this planet and not anywhere else, at least for the moment.
All proteins functions are dependent on their structure, which, in turn, depends on physical and chemical parameters. This is another important fact on studying these molecules; classical biological, physical, chemical, mathematical and informatics sciences have been working together in a new area known as bioinformatics to allow a new level of knowledge about life organization. To understand structural biology, visualization of complex macromolecular structure (like proteins) is essential and macromolecular structure visualization is also now one of the primary steps in the process of drug design and discovery and docking studies. These studies are done virtually thus reducing animal trials and manpower.
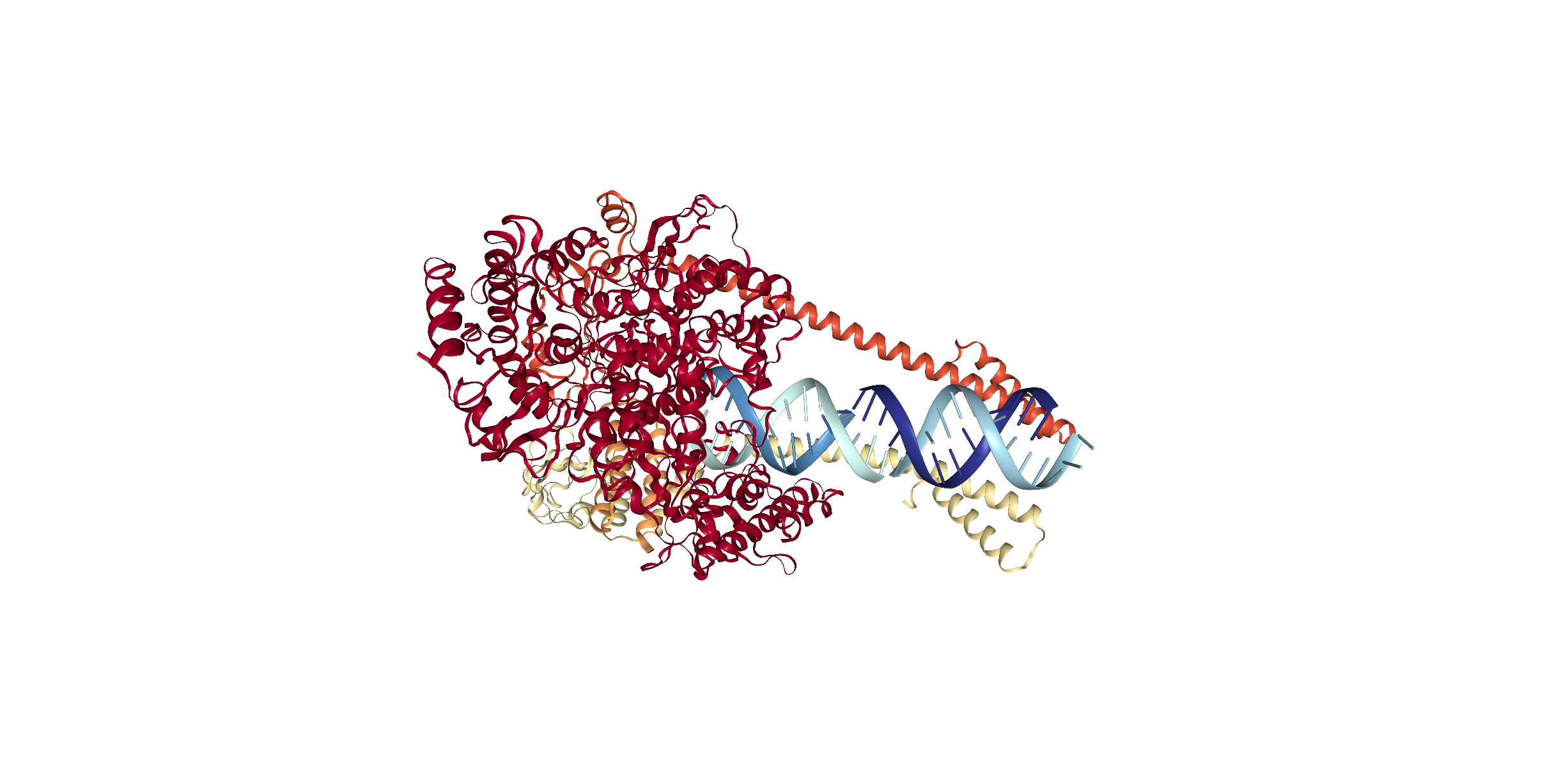
The above visualization depicts the SARS CoV-19 protein 6YYT. This representation is what is called "cartoon format" or "ribbon diagram" and often used as a model for publication purposes. Here, the alpha-helical protein can be described as having many long-range contacts. The two helical stands in blue form the DNA strands of the protein.
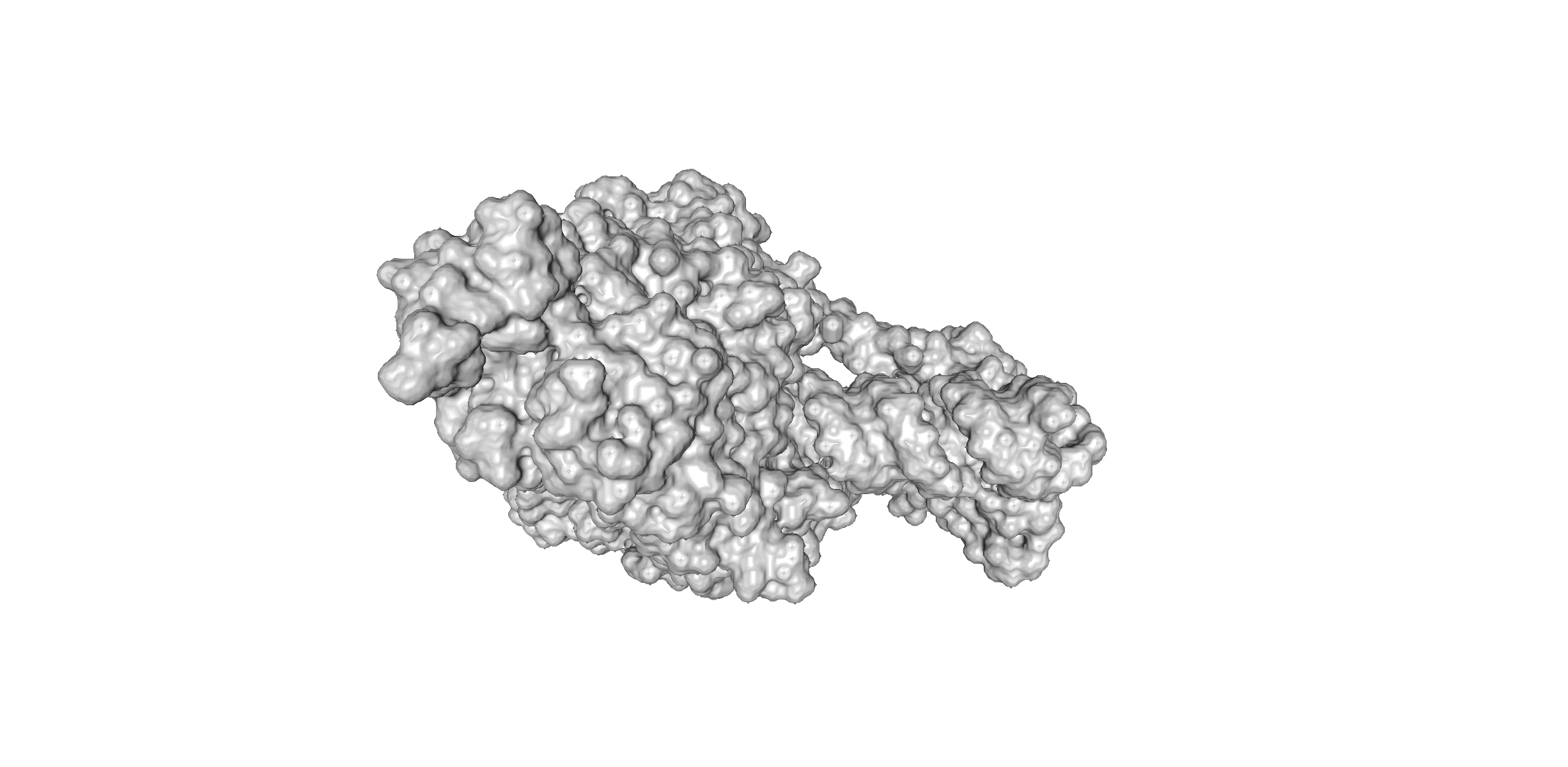
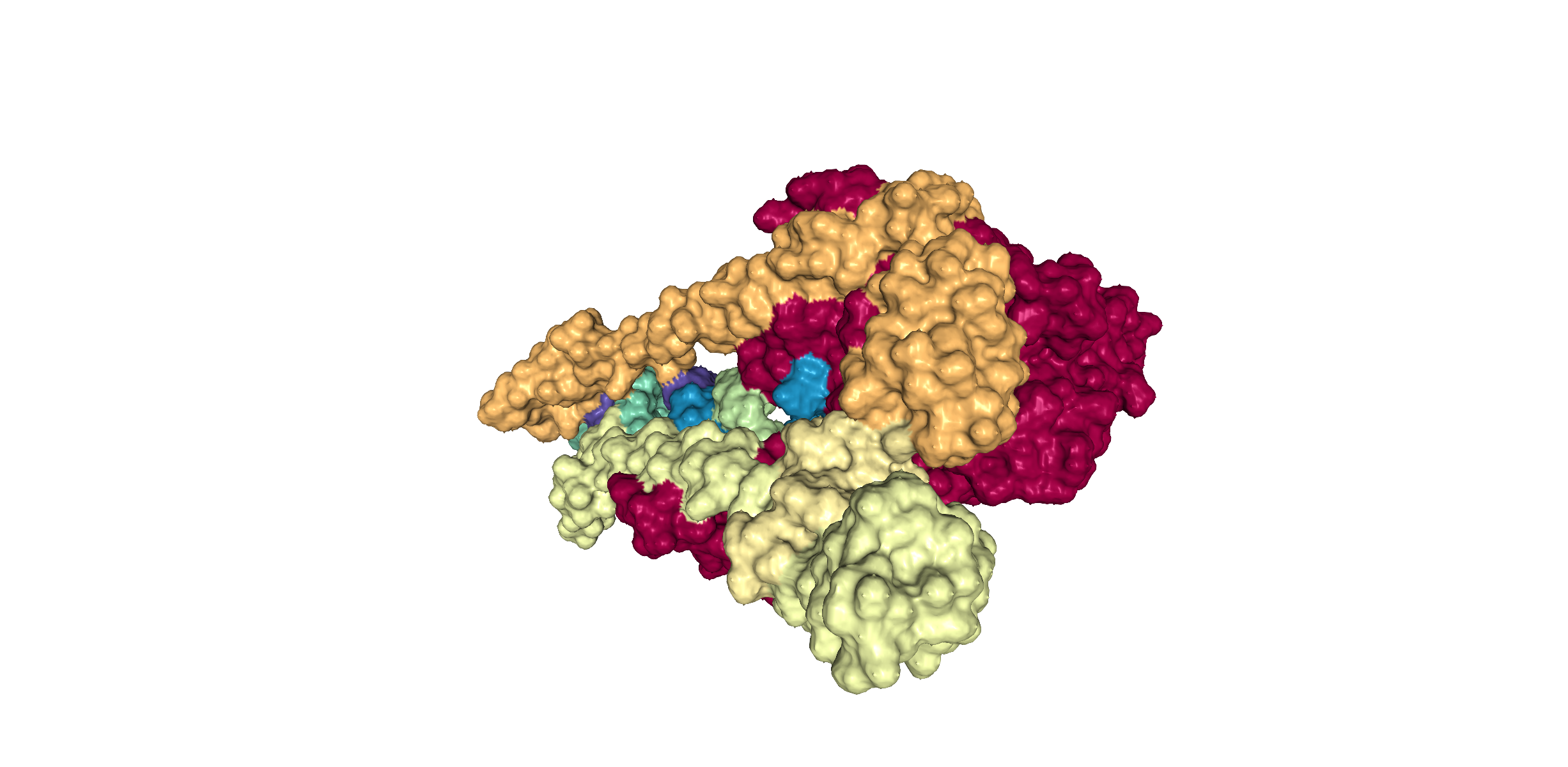
This is the true representation of how the SARS-CoV-19 protein would look like in a biological system. In order for researchers to study complex molecules, the cartoon format of the protein must be converted into what is known as a "surface format" or "surface diagram".
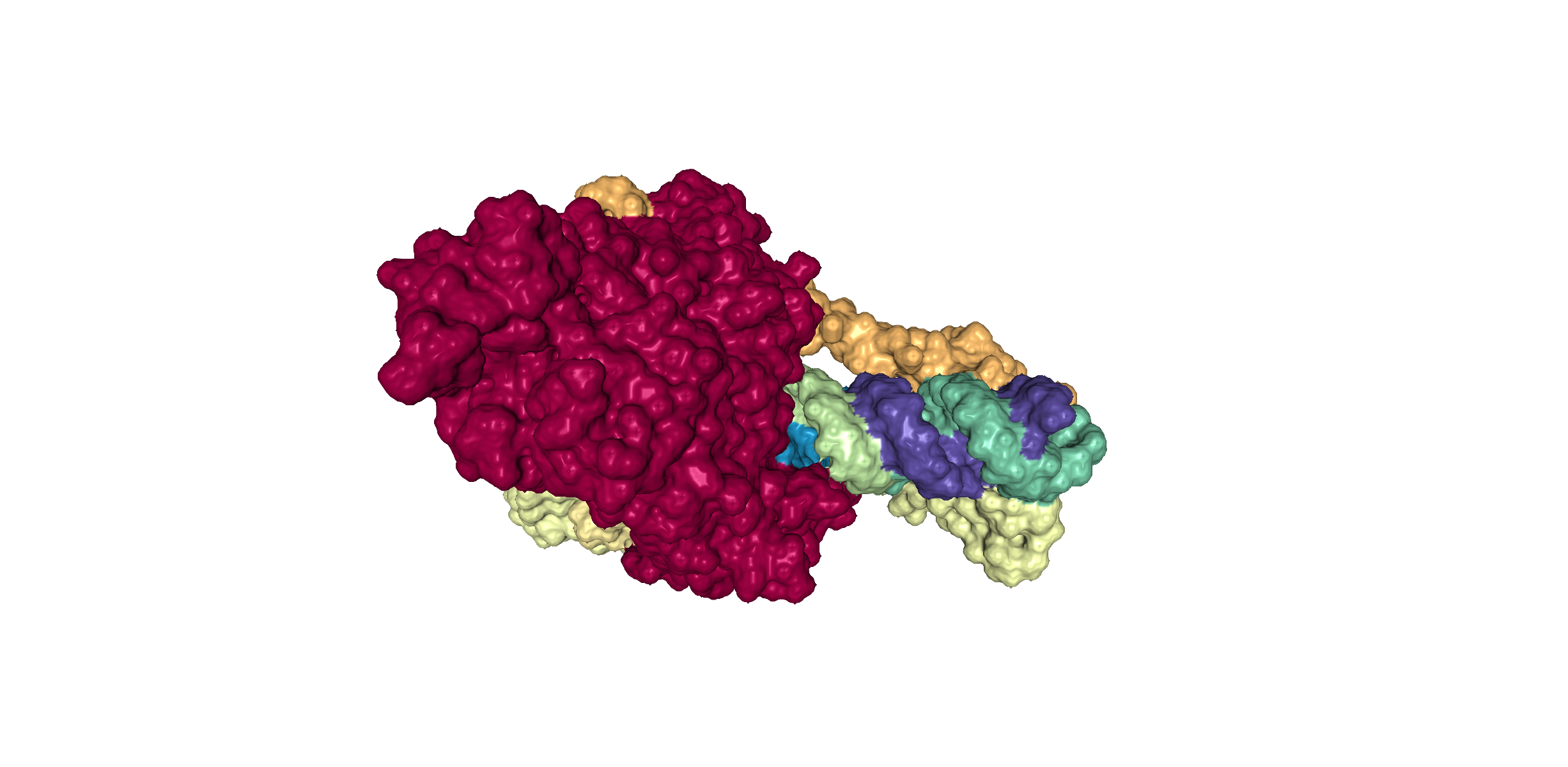
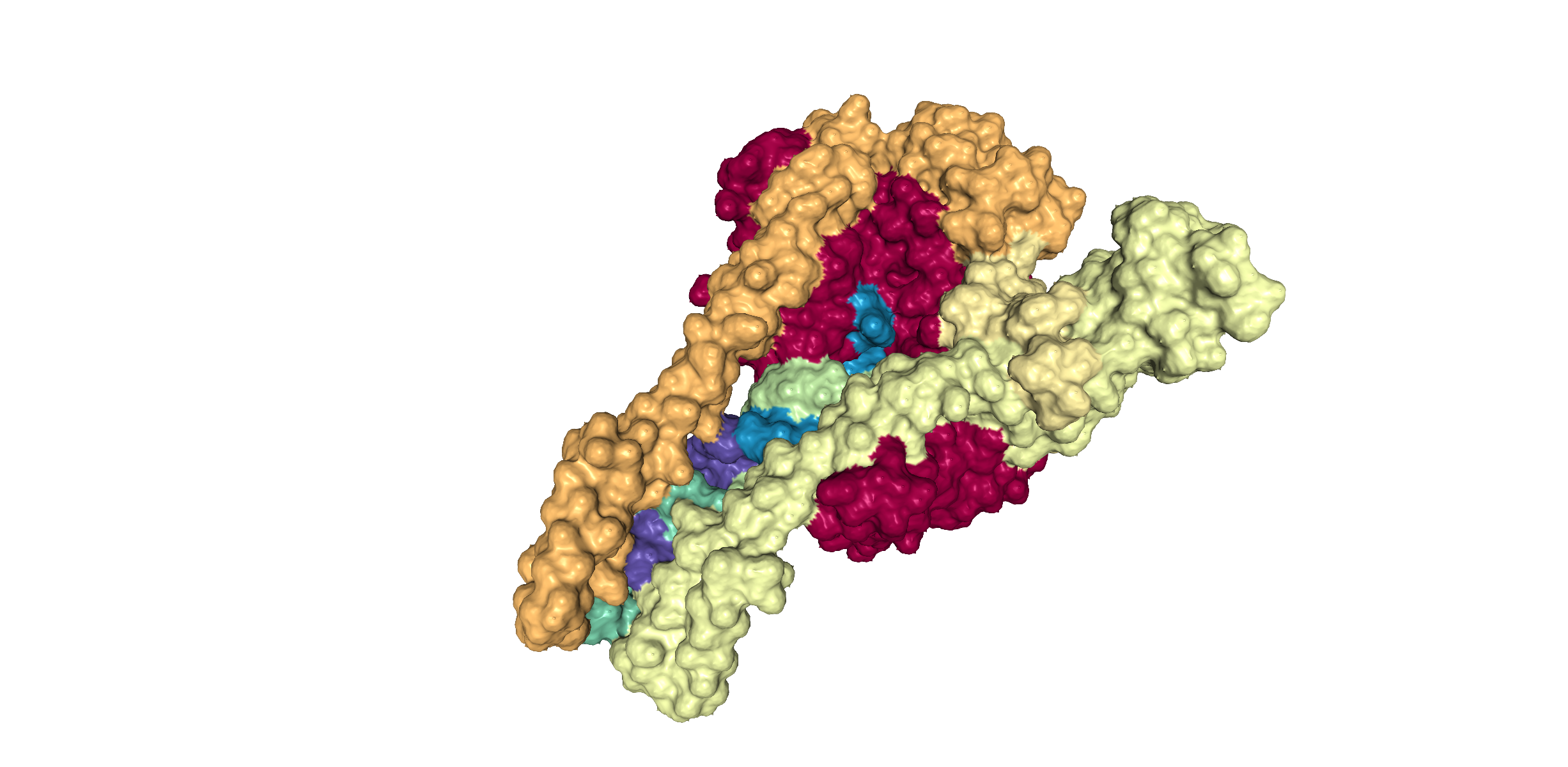
This 3D representation is colored based on the different chains of proteins present in the protein structure. The shading and color adds dimensionality to the diagram. Generally, the features at the front are the highest in contrast and those towards the back are the lowest.
- Sequence length: 29,903 base pairs
- GC content: 37.97%
- Protein content has high Leucine L and Serine S.
- The largest protein is of length 2,701 amino acid.
- Largest protein BLAST results corresponds to SARS-CoV-19 6YYT.
- Protein 6YYT has 8 chains & a RNA binding domain.
- The dataset is available under the Open Database License ODbL.
- Any rights in individual contents of the database are licensed under the Database Contents License.