This aim of this repo is to train a Computer Vision model (CNN, RNN etc.) on MRI scans to identify brain tumor segmentation. A report is available detailing the motivation, research on relevent work and results on model training and tesing.
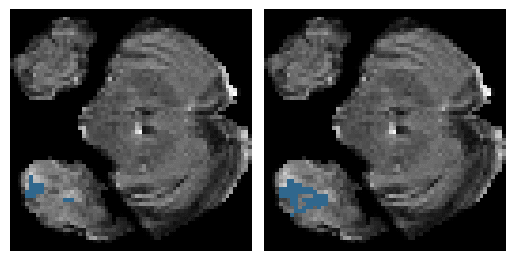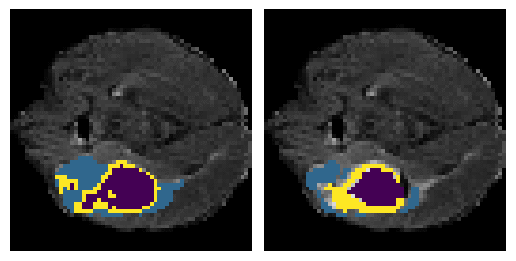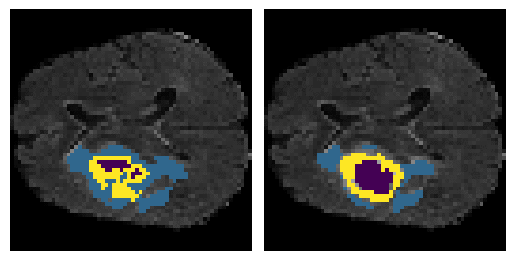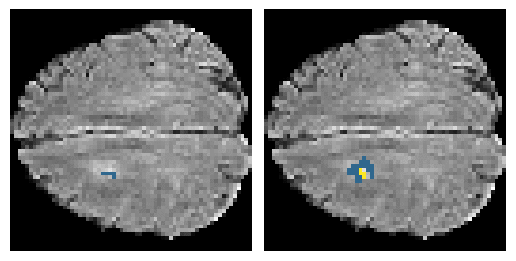
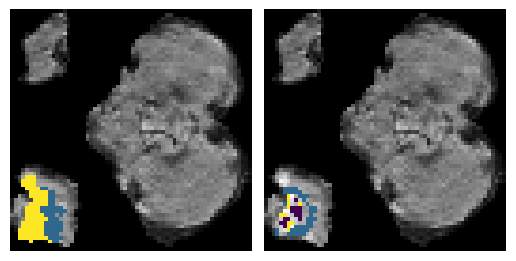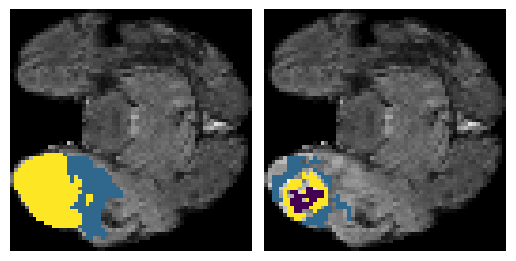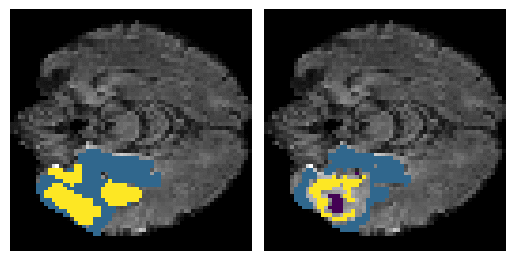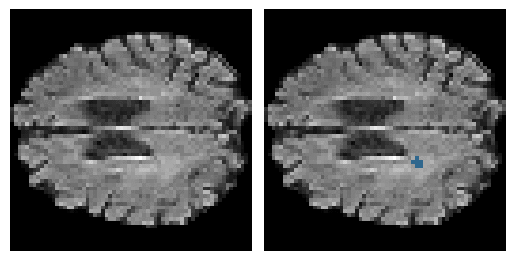
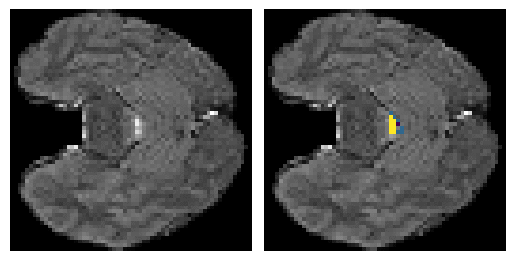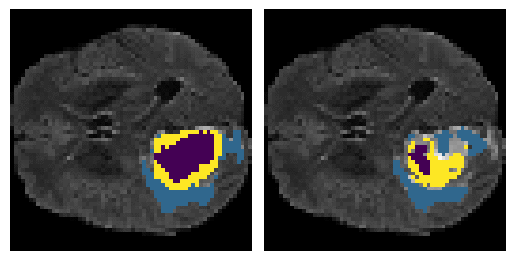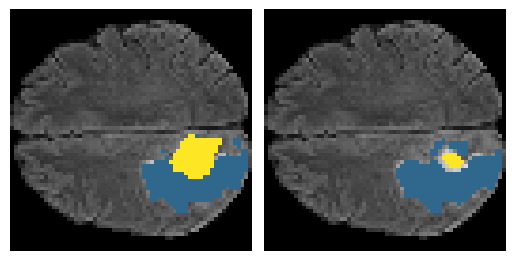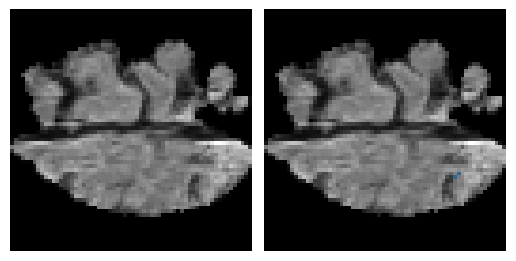
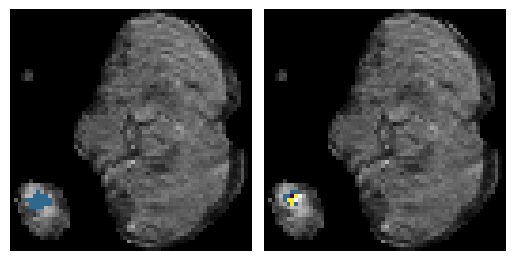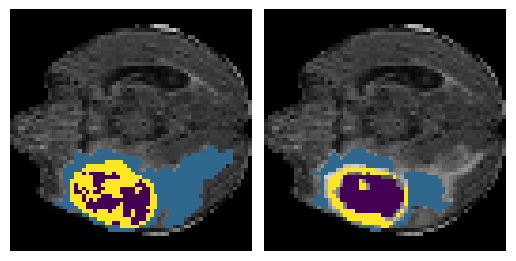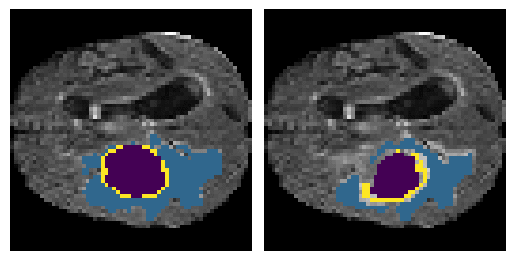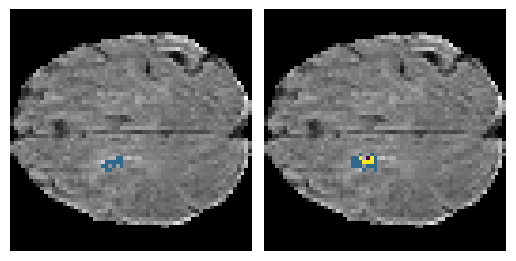
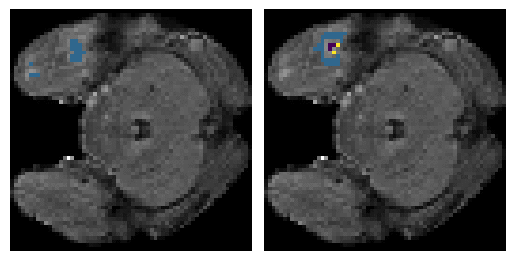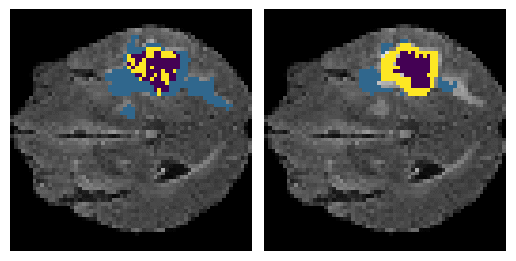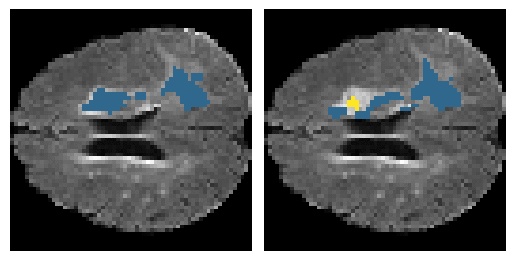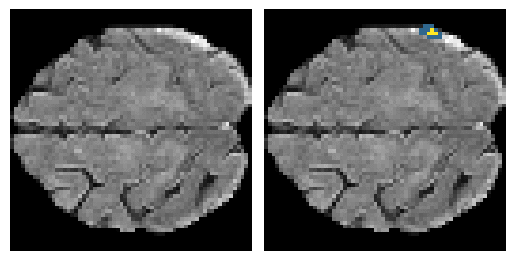
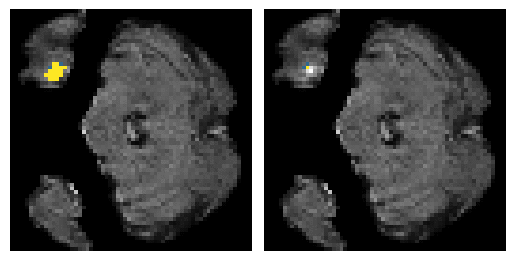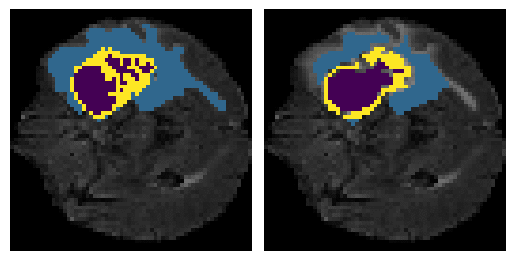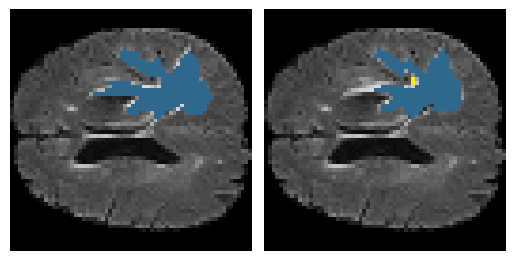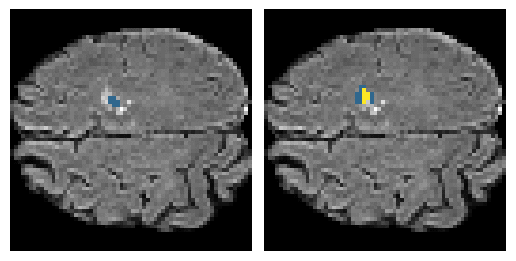
Here is an example of the true data (L) against our predicted result (R). More samples are available at the bottom of this page.
The data is provided by the RSNA-ASNR-MICCAI Brain Tumor Segmentation (BraTS) Challenge 2021 and similar datasets can be downloaded from one of the below:
- www.kaggle.com/datasets/dschettler8845/brats-2021-task1
- www.kaggle.com/competitions/rsna-miccai-brain-tumor-radiogenomic-classification/
- www.kaggle.com/datasets/shakilrana/brats-2023-adult-glioma/
- www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset
All BraTS mpMRI scans are available as NIfTI files (.nii.gz) and describe a) native (T1) and b) post-contrast T1-weighted (T1Gd), c) T2-weighted (T2), and d) T2 Fluid Attenuated Inversion Recovery (T2-FLAIR) volumes, and were acquired with different clinical protocols and various scanners from multiple data contributing institutions.
All the imaging datasets have been annotated manually, by one to four raters, following the same annotation protocol, and their annotations were approved by experienced neuro-radiologists. Annotations comprise the GD-enhancing tumor (ET — label 4), the peritumoral edematous/invaded tissue (ED — label 2), and the necrotic tumor core (NCR — label 1), as described both in the BraTS 2012-2013 TMI paper and in the latest BraTS summarizing paper. The ground truth data were created after their pre-processing, i.e., co-registered to the same anatomical template, interpolated to the same resolution (1 mm3) and skull-stripped.
The model created in this repo contains is a pipeline model containing a Convolutional Autoencoder and a U-Net model. The Convolutional Autoencoder deals with locating the tumour if it was detected by the model, the U-Net model is then used to predict segmentation class given a cropped scan of the tumour. The model inputs are 2D slices of 3D MRI scans, this is done so that the complexity of the model and the number of parameters in the model are drastically reduced. The model is therefore about to predict on brain tumour segmentation on both 2D and 3D MRI scans.
The model training steps can be replicated following the steps shown in 1_train.ipynb. The analysis of the model on the test dataset can be found in 2_test_prediction.ipynb. The code has been tested on Python 3.11.6 and 3.12.0. The list of required libraries and their respective versions can be found in requirements.txt.
6 predictions from the test dataset.
- Left: Ground Truth
- Right: Prediction