This is a python application which takes in an image of a Agar 96 well plate and counts the number of cell colonies. The original application was for automation in synthetic biology and high throughput screening of cell variants. I was tasked with upgrading the accuracy and functionality of a cell colony picking software, who's output gave instructions to a robot which physically picked the cells. I had a lot of trouble finding good open implementations of cell counting, so hopefully this code helps fill that void. After some sleuthing, I adapted the Hough Lines circle counting method to solve the problem, which is significantly better than the legacy code I was replacing (not sure what the algo they used was). If you use this code, please give credit where its due :)
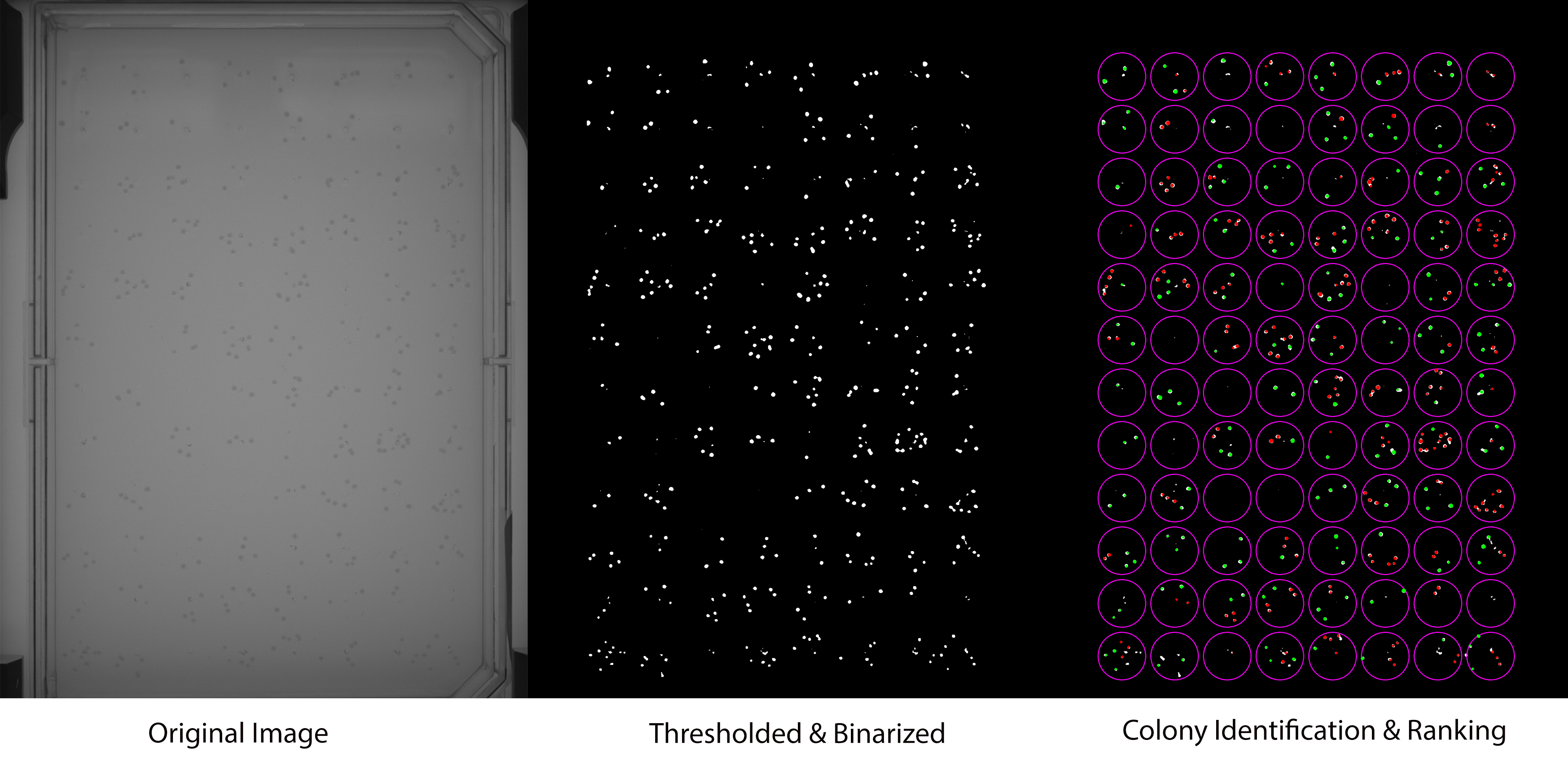
The required setup is a stationary camera and fixed position for the well plate. The algorithm works by subtracting a blank image of the well plate with no cells from the new image in question. This will leave the remaining cells to be counted clearly, albiet with very low contrast. A series of image processing algorithms are applied to boost the features and count the cells via Hough Circles.
The rest of the code and functions serves to pick the "best" colonies for picking by a robot. The criteria for the top ranked cells are the radius of the cell and its separation from other cells. The algorithm will find the top 3 cells in each well plate (determined by preset teach points -purple circles in image) for picking. The green cells above are the "good" cells, and the red ones are deemed non-pickable. It outputs the (x, y) coordinates, radius, and a few other features to a .DAT file for reading by the robot.