The quest to build a fully reproducible software pipeline with provenance
In the quest for truly reproducible workflows I set out to create an example of a reproducible workflow using GNU Guix, IPFS, and CWL. GNU Guix provides content-addressable, reproducible, and verifiable software deployment. IPFS provides content-addressable storage, and CWL describes workflows that can run on specifically supported backend hardware system. In principle, this combination of tools should be enough to provide reproducibility with provenance and improved security.
Note: this work was mostly executed during the Biohackathon 2018 in Matsue, Japan
The Common Workflow Language (CWL) is a specification for describing analysis workflows and tools in a way that makes them portable and scalable across a variety of software and hardware environments——from workstations, to clusters, to the cloud, and to secure high performance computing (HPC) environments. CWL is designed to meet the needs of data-intensive sciences, such as bio- and medial informatics, imaging, astronomy, physics, and chemistry.
CWL started as an answer to problems associated with ad hoc pipeline scripts in bioinformatics, and the acute need to come up with reliable, modular, and well documented solutions. The CWL promises a future of far more efficient pipeline assembly and use, building on components others have created and tested. The objectives of CWL are now more important than ever, especially given the reproducibility crisis. Many important publications can not be reproduced using source data and the reported workflow. These problems are also highlighted in Experimenting with reproducibility: a case study of robustness in bioinformatics and in PiGx: reproducible genomics analysis pipelines with GNU Guix.
CWL definitions are comparatively simple once you understand that responsibilities are split into three components: data inputs (aka Jobs in CWL parlance) are separate from software inputs (aka Tools) and software inputs are split from the workflow definition (fortunately known as Workflows).
The InterPlanetary File System is a protocol and network designed to create content-addressable, peer-to-peer method of storing and sharing hypermedia in a distributed file system. IPFS provides a way of connecting different data sources and sites together (say from different institutes), and can locate and serve files in a (reasonably) scalable way. IPFS is content-addressable (more on this below) and allows for deduplication, local caching, and swarm-like software downloads over the network. IPFS is free software and available through GNU Guix.
GNU Guix is the package manager of the GNU Project. Originally Guix forked the build/install daemon from the Nix project. Guix has morphed into a substantial project in its own right with its own unique packaging system and related tools and with hundreds of committers and roughly 9,000 packages including a wide range of bioinformatics packages. GNU Guix can be deployed on any existing Linux distribution (I use Debian) and provides rigorous and robust controls of dependency graphs.
The short explanation:
Content addressable files are referenced to by a hash on their contents as part of the file path/URI. For example, in the workflow below I use a file named small.chr22.fa that is referenced by its full path:
/ipfs/QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE/small.ERR034597_1.fastq.
A hash value is computed over the fastq file and becomes part of the reference. If the file contents change in any way, even a single letter, the hash value changes, and therefore the reference. This property guarantees you are always dealing with the same input data - a key property of any reproducible pipeline. There can be no ambiguity about file names and what they represent. Files can not change without the file name changing.
Similarly, every GNU Guix software reference includes a hash computed over its inputs which includes the source code and configuration. The reference to a fastq binary executable, for example, looks like
/gnu/store/fijv5bqhf8xmzcys2s70fqvp8xi9vn6m-fastqc-0.11.5/bin/fastqc.A reproducible pipeline therefore includes a unique reference to the binary tool(s). It is even better than that because all dependencies are included in the hash. Therefore the software dependency tree is carved in stone and one can recover and draw the dependency graph as shown below. This may appear a bit involved, but the good news is that most of these references are transparent. The Guix environment deals with resolving them as should become clear.
The first step is to install the Guix. Guix allows regular users to install software packages on any Linux distribution (Debian, Fedora, and CentOS are all fine). GNU Guix does not interfere with running the Linux distribution.
An install script can be run on the command line. That is the easy option. More installation instructions are here at GitLab and here at GNU.org. The short of it is that the Guix (daemon) needs to be installed as root, but runs with user land privileges. For those who can not get access to root there are work arounds, including the use of Docker. Ricardo Wurmus describes how MDC deploys GNU Guix on their HPC and here (essentially use one build host and copy files to the rest). For HPC we typically use a build host that has privileges, but all other HPC nodes simply mount one directory under /gnu/store using a network mount. More HPC blogs on this topic here. If you don’t think it can be done on HPC, think again: Compute Canada deploys Nix on their HPCs (over 120,000 cores). And if you can do Nix, you can do Guix. Same principles.
IPFS was recently added to GNU Guix. The first task for me was to update and add cwltool to GNU Guix. cwltool is the reference implementation of CWL. This took me a few hours because quite a few dependencies had to be added in, and some of these packages have ‘fixated’ versions and ultimately do not build on recent Python 3.7. Of course this should be fixed with cwltools, but with Guix we can introduce both older and recently updated packages without issues, i.e., fixing dependency hell. To manage all this I created a special Guix channel and after setting up the channel (see the README) on Debian, Ubuntu, Fedora, Arch (etc.) the installation should be as easy as
guix package -i cwltool -p ~/opt/cwl
Now to run the tool you need to set the paths etc. with
. ~/opt/cwl/etc/profile cwltool --help
I added the packages in these commits, for example update CWL. Also some packages on Guix trunk needed to be updated, including python-rdflib and python-setuptools. This leads to the following dependency graph for cwltool generated by Guix itself:
(click on the figure to enlarge)
If Guix is correctly installed most packages get downloaded and installed as binaries. Guix only builds packages when it can not find a binary substitute. And now I can run
cwltool --version /gnu/store/nwrvpgf3l2d5pccg997cfjq2zqj0ja0j-cwltool-1.0.20181012180214/bin/.cwltool-real 1.0
Success!
Note that the guix-cwl channel also provides a Docker image that we will update for cwltool.
After adding the cwl channel we can have the main tools installed in one go with
guix package -i go-ipfs cwltool -p ~/opt/cwlAgain, to make the full environment available do
. ~/opt/cwl/etc/profile
ipfs --version
ipfs version 0.4.19First, I thought to run one of the pipelines from bcbio-nextgen as an example. Bcbio conveniently generates CWL (they switched from Python scripts to CWL scripts). But then at the BH18 there was a newly created CWL pipeline in https://github.com/hacchy1983/CWL-workflows and I decided to start from there. This particular pipeline uses github to store data and a Docker container to run a JVM tool. Good challenge to replace that with IPFS and Guix and make it fully reproducible.
Note that git does provide provenance but is not suitable for large data files. And even though Docker may provide reproducible binary blobs, it is quite hard to verify what is in them, i.e., there is a trust issue, and it is usually impossible to recreate them exactly——the core of the reproducibility issue. We can do better than this.
In the next step we are going to make the data available through IPFS (as installed above).
After the installation of go-ipfs, create a data structure following the IPFS instructions directory
mkdir /export/data/ipfs
env IPFS_PATH=/export/data/ipfs ipfs init
initializing IPFS node at /export/data/ipfs
generating 2048-bit RSA keypair...done
peer identity: QmUZsWGgHmJdG2pKK52eF9kG3DQ91fHWNJXUP9fTbzdJFRStart the daemon
env IPFS_PATH=/export/data/ipfs ipfs daemon(note that ipfs uses quite a bit of bandwidth to talk to its peers. For that reason don’t keep the daemon running on a mobile network, for example).
And now we can add the data
export IPFS_PATH=/export/data/ipfs
ipfs add -r DATA2/
added QmXwNNBT4SyWGnNogzDq8PTbtFi48Q9J6kXRWTRQGmgoNz DATA/small.ERR034597_1.fastq
added QmcJ7P7eyMqhttSVssYhiRPUc9PxqAapVvS91Qo78xDjj3 DATA/small.ERR034597_2.fastq
added QmfRb8TLfVnMbxauTPV2hx5EW6pYYYrCRmexcYCQyQpZjV DATA/small.chr22.fa
added QmXaN36yNT82jQbUf2YuyV8symuF5NrdBX2hxz4mAG1Fby DATA/small.chr22.fa.amb
added QmVM3SERieRzAdRMxpLuEKMuWT6cYkhCJsyqpGLj7qayoc DATA/small.chr22.fa.ann
added QmfYpScLAEBXxyZmASWLJQMZU2Ze9UkV919jptGf4qm5EC DATA/small.chr22.fa.bwt
added Qmc2P19eV77CspK8W1JZ7Y6fs2xRxh1khMsqMdfsPo1a7o DATA/small.chr22.fa.pac
added QmV8xAwugh2Y35U3tzheZoywjXT1Kej2HBaJK1gXz8GycD DATA/small.chr22.fa.sa
added QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE DATATest a file
ipfs cat QmfRb8TLfVnMbxauTPV2hx5EW6pYYYrCRmexcYCQyQpZjVand you should see the contents of small.chr22.fa. You can also browse to http://localhost:8080/ipfs/QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE on your local machine.
Next you ought to pin the data so it does not get garbage collected by IPFS.
ipfs pin add QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE
pinned QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE recursivelyFollow the instructions in the original workflow README
cwltool Workflows/test-workflow.cwl Jobs/small.ERR034597.test-workflow.ymlwhere the first CWL describes the workflow and the second the data inputs. This command complains we don’t have Docker. Since we want to run without Docker specify –no-container:
cwltool --no-container Workflows/test-workflow.cwl Jobs/small.ERR034597.test-workflow.ymlResulting in
'fastqc' not found: [Errno 2] No such file or directory: 'fastqc': 'fastqc'
which exists in Guix, so
guix package -i fastqc -p ~/opt/cwlinstalls
fastqc 0.11.5 /gnu/store/sh0wj2c00vkkh218jb5p34gndfdmbhrf-fastqc-0.11.5
and also downloads missing fastqc dependencies
/gnu/store/sh0wj2c00vkkh218jb5p34gndfdmbhrf-fastqc-0.11.5
/gnu/store/0j2j0i55s0xykfcgx9fswks8792gk4sk-java-cisd-jhdf5-14.12.6-39162
/gnu/store/bn8vb4zvdxpjl6z573bxyzqndd925x97-java-picard-1.113
/gnu/store/g08d57f1pbi6rrzlmcaib1iyc6ir5wn9-icedtea-3.7.0
/gnu/store/m0k3fdpgyms3fwbz24vaxclx6f1rwjdg-java-jbzip2-0.9.1Note that the package is completely defined with its dependencies and ‘content-addressable’. We can see it pulls in Java and Picard. Note also the software is made available under an ‘isolated’ profile in ~/opt/cwl. We are not mixing with other software setups. And, in the end, all software installed in this profile can be hosted in a (Docker) container.
After installing with Guix we can rerun the workflow and note that it fails at the next step with
/gnu/store/nwrvpgf3l2d5pccg997cfjq2zqj0ja0j-cwltool-1.0.20181012180214/bin/.cwltool-real 1.0
Resolved 'Workflows/test-workflow.cwl' to '/hacchy1983-CWL-workflows/Workflows/test-workflow.cwl'
[workflow ] start
[workflow ] starting step qc1
[step qc1] start
[job qc1] /tmp/ig4k8x8m$ fastqc \
-o \
. \
/tmp/tmp0m1p3syh/stgca222f81-6346-4abf-a005-964e80dcf783/small.ERR034597_1.fastq
Started analysis of small.ERR034597_1.fastq
Approx 5% complete for small.ERR034597_1.fastq
Approx 10% complete for small.ERR034597_1.fastq
Approx 15% complete for small.ERR034597_1.fastq
Approx 20% complete for small.ERR034597_1.fastq
...
Error: Unable to access jarfile /usr/local/share/trimmomatic/trimmomatic.jarPartial success. fastqc runs fine and now we hit the next issue. The /usr/local points out there is at least one problem :). There is also another issue in that the data files are specified from the source tree, e.g.
fq1: # type "File"
class: File
path: ../DATA/small.ERR034597_1.fastq
format: http://edamontology.org/format_1930Here you may start to appreciate the added value of a CWL workflow definition. By using an EDAM ontology CWL gets metadata describing the data format which can be used down the line.
To make sure we do not fetch the old data I moved the old files out of the way and modified the job description to use the IPFS local web server
git mv ./DATA ./DATA2
mkdir DATAWe need to fetch with IPFS so the description becomes
--- a/Jobs/small.ERR034597.test-workflow.yml
+++ b/Jobs/small.ERR034597.test-workflow.yml
@@ -1,10 +1,10 @@
fq1: # type "File"
class: File
- path: ../DATA/small.ERR034597_1.fastq
+ path: http://localhost:8080/ipfs/QmR8..h1tE/small.ERR034597_1.fastq
format: http://edamontology.org/format_1930
fq2: # type "File"
class: File
- path: ../DATA/small.ERR034597_2.fastq
+ path: http://localhost:8080/ipfs/QmR8..h1tE/small.ERR034597_2.fastq
format: http://edamontology.org/format_1930
fadir: # type "Directory"
class: DirectoryThe http fetches can be replaced later with a direct IPFS call which
will fetch files transparently from the public IPFS somewhere - much
like bit torrent does - and cache locally. We will need to add that
support to cwltools (which can possibly be done as a plugin) so we can write
something like
path: ipfs://QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE
This would be safe because IPFS is content-addressable.
Now the directory tree looks like
tree
.
├── DATA
├── DATA2
│ ├── small.chr22.fa
│ ├── small.chr22.fa.amb
│ ├── small.chr22.fa.ann
│ ├── small.chr22.fa.bwt
│ ├── small.chr22.fa.pac
│ ├── small.chr22.fa.sa
│ ├── small.ERR034597_1.fastq
│ └── small.ERR034597_2.fastq
├── Jobs
│ ├── small.chr22.bwa-index.yml
│ └── small.ERR034597.test-workflow.yml
├── LICENSE
├── README.md
├── small.ERR034597_1_fastqc.html
├── Tools
│ ├── bwa-index.cwl
│ ├── bwa-mem-PE.cwl
│ ├── fastqc.cwl
│ ├── samtools-sam2bam.cwl
│ └── trimmomaticPE.cwl
└── Workflows
└── test-workflow.cwland again CWL runs up to
ILLUMINACLIP:/usr/local/share/trimmomatic/adapters/TruSeq2-PE.fa:2:40:15 Error: Unable to access jarfile /usr/local/share/trimmomatic/trimmomatic.jar
The original workflow pulls trimmomatic.jar as a Docker image. Just as an example here I download the jar file and created a GNU Guix package to make it available to the workflow.
Guix likes things to be built from source. This is a clear goal of the GNU project. But you can still stick in binary blobs if you want. Main thing is that they need to be available in the /gnu/store to be seen at build/install time. Here I am going to show you how to do this, but keep in mind that for reproducible pipelines this is a questionable design choice.
I created a jar download for GNU Guix. This was done by creating a Guix channel as part of the repository. The idea of the package in words is:
- Download the jar and compute the HASH for Guix with
guix download http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic/Trimmomatic-0.38.zip
/gnu/store/pkjlw42f5ihbvx2af6macinf290l3197-Trimmomatic-0.38.zip
0z34y7f9idnxgnyqdc29z4hwdp8f96mlqssyxvks4064nr1aya6l- Check the contents of the Zip file
unzip -t /gnu/store/pkjlw42f5ihbvx2af6macinf290l3197-Trimmomatic-0.38.zip
testing: Trimmomatic-0.38/trimmomatic-0.38.jar OK- On running ‘guix install’ Guix will unzip the file in a ‘build’ directory
- You need to tell Guix to copy the file into the target ‘installation’ directory -
we’ll copy it into
lib/share/jar - After installation the jar will be available in the profile under that directory path
A (paraphrased) YAML definition therefore looks like:
- fetch:
url: http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic/Trimmomatic-0.38.zip
hash: 0z34y7f9idnxgnyqdc29z4hwdp8f96mlqssyxvks4064nr1aya6l
- dependencies:
- java
- unzip
- build:
- unzip zipfile
- copy-recursively "Trimmomatic-0.38" to targetIf you want to see the actual package definition and how it is done
see
https://github.com/pjotrp/CWL-workflows/blob/0f1c3c971f19956ca445a4ba50f575e972e4e835/package/trimmomatic.scm. The
package is written in Scheme, and if you think away the parenthesis
you have pretty much what I have described. Note that one advantage of
using Scheme is that we can define inline variables, such as source
and target. Something CWL does by including a full blown Javascript
interpreter.
After installing the package and updating the profile, try again after updating the paths for trimmomatic in
env GUIX_PACKAGE_PATH=../hacchy1983-CWL-workflows/ \
./pre-inst-env guix package -i trimmomatic-jar -p ~/opt/cwl
# ---- Update the paths
. ~/opt/cwl/etc/profile
# ---- Run
cwltool --no-container Workflows/test-workflow.cwl Jobs/small.ERR034597.test-workflow.ymlThe GUIX_PACKAGE_PATH points into the workflow directory where I created the package.
In the next step the workflow failed because bwa was missing, so added bwa with Guix
guix package -i bwa -p ~/opt/cwlAnd then we got a different error
[E::bwa_idx_load_from_disk] fail to locate the index files
This workflow is broken because there are no index files! The original CWL script
just assumed they were in the fadir directory.
If you check the earlier IPFS upload you can see we added them with:
added QmfRb8TLfVnMbxauTPV2hx5EW6pYYYrCRmexcYCQyQpZjV DATA/small.chr22.fa
added QmXaN36yNT82jQbUf2YuyV8symuF5NrdBX2hxz4mAG1Fby DATA/small.chr22.fa.amb
added QmVM3SERieRzAdRMxpLuEKMuWT6cYkhCJsyqpGLj7qayoc DATA/small.chr22.fa.ann
added QmfYpScLAEBXxyZmASWLJQMZU2Ze9UkV919jptGf4qm5EC DATA/small.chr22.fa.bwt
added Qmc2P19eV77CspK8W1JZ7Y6fs2xRxh1khMsqMdfsPo1a7o DATA/small.chr22.fa.pac
added QmV8xAwugh2Y35U3tzheZoywjXT1Kej2HBaJK1gXz8GycD DATA/small.chr22.fa.sa
added QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE DATABut the workflow does not automatically fetch them. So, we need to fix that. Just add them using IPFS (though we could actually recreate them using ‘bwa index’ instead).
diff --git a/Jobs/small.ERR034597.test-workflow.yml b/Jobs/small.ERR034597.test-workflow.yml
index 9b9b153..51f2174 100644
--- a/Jobs/small.ERR034597.test-workflow.yml
+++ b/Jobs/small.ERR034597.test-workflow.yml
@@ -6,7 +6,18 @@ fq2: # type "File"
class: File
path: http://localhost:8080/ipfs/QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE/small.ERR034597_2.fastq
format: http://edamontology.org/format_1930
-fadir: # type "Directory"
- class: Directory
- path: ../DATA
-ref: small.chr22 # type "string"
+ref: # type "File"
+ class: File
+ path: http://localhost:8080/ipfs/QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE/small.chr22.fa
+ format: http://edamontology.org/format_1929
+ secondaryFiles:
+ - class: File
+ path: http://localhost:8080/ipfs/QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE/small.chr22.fa.amb
+ - class: File
+ path: http://localhost:8080/ipfs/QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE/small.chr22.fa.ann
+ - class: File
+ path: http://localhost:8080/ipfs/QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE/small.chr22.fa.bwt
+ - class: File
+ path: http://localhost:8080/ipfs/QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE/small.chr22.fa.pac
+ - class: File
+ path: http://localhost:8080/ipfs/QmR81HRaDDvvEjnhrq5ftMF1KRtqp8MiwGzwZnsC4ch1tE/small.chr22.fa.saTo make the workflow work I had to replace the concept of an fa directory for bwa to using these files explicitly which better describes what is happening (as a bonus):
diff --git a/Tools/bwa-mem-PE.cwl b/Tools/bwa-mem-PE.cwl
index fc0d12d..0f87af3 100644
--- a/Tools/bwa-mem-PE.cwl
+++ b/Tools/bwa-mem-PE.cwl
@@ -19,12 +19,17 @@ requirements:
baseCommand: [ bwa, mem ]
inputs:
- - id: fadir
- type: Directory
- doc: directory containing FastA file and index
- id: ref
- type: string
- doc: name of reference (e.g., hs37d5)
+ type: File
+ inputBinding:
+ position: 2
+ doc: Fasta reference (e.g., hs37d5)
+ secondaryFiles:
+ - .amb
+ - .ann
+ - .bwt
+ - .pac
+ - .sa
- id: fq1
type: File
format: edam:format_1930After that we got
Final process status is success
Yes!
The source and full diff can be viewed on github.
GNU Guix has an option to rebuild packages multiple times and compare the results. In case there is a difference the packages can not be considered deterministic. For example software builds may contain a time stamp at time of build. This is harmless, but who is to tell the difference is not caused by something else? This is why the reproducible builds project exist of which Guix is a member. See also GNU Guix Reproducible builds: a means to an end.
The referenc CWL runner does not have such an option (yet). I ran it by hand three times. The first time capture the MD5 values with
find . -type f -print0 | xargs -0 md5sum > ~/md5sum.txt
next times check with
md5sum -c ~/md5sum.txt |grep -v OK
it complained on one file
./output.sam: FAILED md5sum: WARNING: 1 computed checksum did NOT match
and the @PG field in the output file contains a temporary path:
diff output.sam output.sam.2
2c2
< @PG ID:bwa PN:bwa VN:0.7.17-r1188 CL:bwa mem -t 4 /gnu/tmp/cwl/tmpdoetk_3r/stge19b3f1c-864a-478e-8aee-087a61654aba/small.chr22.fa /gnu/tmp/cwl/tmpdoetk_3r/stgd649e430-caa8-491f-8621-6a2d6c67dcb9/small.ERR034597_1.fastq.trim.1P.fastq /gnu/tmp/cwl/tmpdoetk_3r/stg8330a0f5-751e-4685-911e-52a5c93ecded/small.ERR034597_2.fastq.trim.2P.fastq
---
> @PG ID:bwa PN:bwa VN:0.7.17-r1188 CL:bwa mem -t 4 /gnu/tmp/cwl/tmpl860q0ng/stg2210ff0e-184d-47cb-bba3-36f48365ec27/small.chr22.fa /gnu/tmp/cwl/tmpl860q0ng/stgb694ec99-50fe-4aa6-bba4-37fa72ea7030/small.ERR034597_1.fastq.trim.1P.fastq /gnu/tmp/cwl/tmpl860q0ng/stgf3ace0cb-eb2d-4250-b8b7-eb79448a374f/small.ERR034597_2.fastq.trim.2P.fastqTo fix this we could add a step to the pipeline to filter out this field or force output to go into the same destination directory. Or tell bwa to skip the @PG field.
Determinism (and reproducibility) may break when the pipeline has software that does not behave well. Some tools give different results when run using identical inputs. Unfortunately, the solution is to fix or avoid such software. Also, software may try to download inputs which can lead to different results over time, for example by including a time stamp in the output. To be stringent, it may be advisable to disable network traffic when the workflow is running. GNU Guix builds all its software without a network, i.e., after downloading the files as described in the package definition the network is switched off and the build procedure runs without network in complete isolation. This guarantees software can not download non-deterministic material from the internet. It also guarantees no dependencies can ‘bleed’ in. This is why GNU Guix is called a ‘functional package manager’ - in the spirit of functional programming.
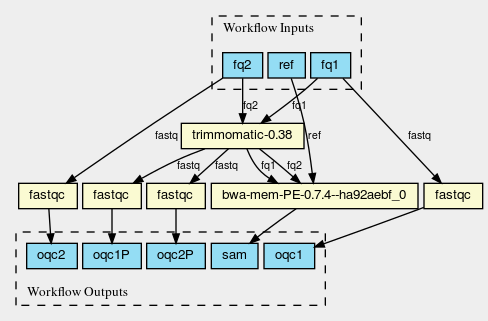
This figure shows the dependency graph for running the workflow, and includes fastqc, trimmomatic-jar, bwa, ipfs-go, and cwltool itself.
(click on the figure to enlarge)
This is a huge graph (but not abnormal). GNU Guix keeps track of all these dependencies (here we show versions, but can also show the hash values) and can therefore easily display the current graph. Note that the full graph that includes all build dependencies to create the software is much larger.
The trend is that most software depends on an increasing number of other software compilers, tools, libraries, and modules. To remain stress-free and sane, a rigorous way of managing resources is crucial and this is what GNU Guix provides.
The figure was created by adding the workflow to the CWL viewer online (simply by pasting the github link). See https://view.commonwl.org/workflows/github.com/pjotrp/CWL-workflows/blob/guix-cwl/Workflows/test-workflow.cwl
There are two issues with the cwlviewer. First, the PNG/SVG output links do not end in .png and .svg respectively. This makes it hard to show them in a browser. Second, I could not find out how to update a view once a repo had been linked. It should be possible to update information - at least to create a new generation of workflow. I have now been informed you can only update after 24 hours… That is an odd restriction.
Now you may ask at this point: what is actually the difference with the original workflow? There are a few differences - first we were forced to make the inputs more explicit. In the original there was no mention of BWA index files, they just sat in the github repository. The main difference, however, is that we were forced to specify all tools and their dependencies. The original workflow simply assumed the tools would already be on the system including the CWL runner cwltool itself! The tools were specified as CWL hints:
hints:
- class: DockerRequirement
dockerPull: 'quay.io/biocontainers/fastqc:0.11.7--pl5.22.0_2'The Docker link is a ‘hint’ which means the CWL runner will try to fetch the image using Docker by default. Without enabling Docker (the –no-container switch), local installations of tools get preferential treatment. So, apart from downloading a separate Docker image for every tool (and every time on a HPC compute node) we also have the risk of tools ‘bleeding’ in from the local environment, depending on how the CWL runner is behaving/deployed. This is because software paths are not rigourously fixated in CWL scripts - it has to be handled by then environment. One way to handle this is by using a GNU Guix container.
To fixate dependencies AND to ascertain no tools bleed in to our workflow from the underlying system AND to make sure we don’t miss out on any dependencies we can run our workflow inside a GNU Guix container. This is not a Docker container - more on that in the next section. Let’s create a container.
The original command was
env TMPDIR=/gnu/tmp/cwl cwltool --preserve-environment TMPDIR \
--preserve-environment GUIX_PROFILE --leave-tmpdir \
--no-container Workflows/test-workflow.cwl Jobs/small.ERR034597.test-workflow.ymlNow we are going to run that inside a Guix container this means only the items that are dependencies of the tools we specify are included in the container. Note that we switch on networking to be able to fetch data through IPFS:
env GUIX_PACKAGE_PATH=../hacchy1983-CWL-workflows \
guix environment --network -C guix \
--ad-hoc cwltool trimmomatic-jar bwa fastqc go-ipfs curlNow run the workflow with
cwltool --no-container Workflows/test-workflow.cwl Jobs/small.ERR034597.test-workflow.ymlI first had to update the Guix profile so as to use the direct store path in the new container for trimmomatic - but otherwise it works as advertised. See the output (the listed error is harmless, but I should fix it).
Now we have the software stack in a GNU Guix container we can also have Guix create a Docker container with
guix pack -f docker cwltool trimmomatic-jar bwa fastqc go-ipfs
/gnu/store/57fg8hfah46rclg3vybb9nckg6766izp-docker-pack.tar.gzwhich writes out a container that can be uploaded to docker hub or some other repo without using Docker. See also https://github.com/genenetwork/guix-cwl where we dit exactly that. A recent version of Docker is packaged in GNU Guix.
Let’s be optimisitic and assume we have all software running correctly in an isolated container created by GNU Guix and we have fetched all data as inputs from IPFS. We will then have achieved a fully reproducible pipeline that can be uploaded on the internet and then be run by anyone anywhere.
There are two improvements to be made:
- Include the CWL scripts in the container
- Create a package definitions that forces the dependencies for
cwltool trimmomatic-jar bwa fastqc go-ipfsinto the container so we can do
guix pack -f docker my-workflowAnd everything is pulled into the container. We could even make a Guix package (and therefor container) that includes all data inputs.
I will leave this as an exercise for the reader right now, but with research objects and ‘live publications’ the enforcement of such practices may be around the corner.
In the near future we can imagine that a scientific publication is accompanied by a ‘live document’. I.e., the pipeline with datasets can be rerun by anyone, anywhere. And results can be reproduced and validated. With the current technology stack it can become a common requirement with journal publications. Prototypes of such live publications should appear in the coming two years.
Here I have explained some of the principle and mechanics of building a reproducible pipeline. With little effort, anyone should be able create such a pipeline using GNU Guix, an addressable data source such as IPFS, and a CWL work flow definition that includes content-addressable references to software and data inputs (here I used IPFS for data). By running the workflow multiple times we can asserted that the outcome is deterministic (save for hardware failure, cosmic rays, acts-of-god, and super villains) and therefore reproducible.
In the process of migrating the original Docker version of this workflow it became evident that not all inputs were explicitly defined.
This reproducible workflow captures the full graph, including all data, tools, and the cwl-runner itself! There was no need to use Docker at all. In fact, this version is better than the original Docker pipeline because both software and data are complete, and are guaranteed to run with the same (binary) tools.
To guarantee reproducibility it is necessary to fixate the inputs and have well behaved software. With rogue or badly behaved software this may be a challenge. The good news is that such behaviour is not so common and when encountered, GNU Guix and IPFS will highlight the reproducibility problems.
CWL includes a range of satellite tools including cwlviewer that I used to generate the workflow information. Such tools come for ‘free’ with the CWL. CWLviewer is useful for discovering workflows created by other researchers and to find examples of CWL scripts.
The bottom line here is that CWL is a very powerful technical solution for generating pipelines that can be shared. It is reasonably simple, and responsibilities are split into three managable pieces. Data inputs are separate from software inputs and software inputs are separated from workflow definitions. The online documentation for CWL is still sparse. For example, to figure out the use of secondaryFiles for bwa I read through a number of existing pipelines on Github. But with the growth of online pipelines, CWL will become stronger and stronger. With this growing support any CWL user will gain the benefit of capturing provenance graphs and other goodies.
Beside improving the documentation, I suggest CWL runners add an option for checking determinism (run workflows multiple times and check results), add support for native IPFS (a Python IPFS implementation exists, alternatively IPFS fuse could be used) and add some support for GNU Guix profiles - one single variable pointing in the GUIX_PROFILE path - so it becomes even easier to create deterministic software deployments that are built from source, transparent and recreatable for eternity (which is a very long time).
Docker has had a good run over the last few years, but with respect to these last two points – transparency and recreatability – Docker really falls short. A Docker image is a binary ‘blob’ and it is impossible to go back from the image alone and see how it was built. This is not transparent. In almost all cases, Docker build instructions include the equivalent of a ‘apt-get update’ which essentially says the image will end up being different every time you create it. This means it is virtually impossible to recreate an image. The greatest concern, however, is that of trust. Downloading a binary blob over the internet is not a great idea and especially when dealing with privacy concerns.
GNU Guix provides a viable alternative in that (1) it is built from source which means a workflow with tools can be audited and considered more secure and (2) provides full transparency, recreatability (read faithful reproducibility). With GNU Guix and CWL you don’t need Docker, though it is still possible to run Guix created Docker images that also have these Guix advantages. Another advantage of Guix containers over Docker containers is that they are lighter and therefore faster.
Finally, we are working on a workflow language that integrates reproducible software deployment: the Guix Workflow Language, or GWL. This is still work-in-progress but we believe this approach is promising because it is both simpler and more rigorous and can be combined with CWL, and in the future it may write CWL definitions. I am sure I’ll introduce a mix of GWL and CWL workflows in my pipelines in the near future. Guix could also benefit from an online repository of channels and pipelines similar to view.commonwl.org. Very useful. Kudo’s to the creators.
The full package graph can be generated with
guix graph cwltool |dot -Tpdf > cwltool-package.pdfWe also create a graph for all tools in this workflow we can do
guix graph cwltool go-ipfs trimmomatic-jar bwa fastqc | dot -Tpdf > full.pdfAnd the full dependency graph for cwltool, that includes the build environment, can be generated with
guix graph --type=references cwltool |dot -Tpdf > cwltool-references.pdfguix pack -f docker cwltool trimmomatic-jar bwa fastqc go-ipfs curl