Repository to share data, workflows, apps, and scripts working toward the goal of execution of BioCompute Objects (BCO).
This repository also includes two example workflows that can be used to generate a BCO. One, is a simple CWL tool definition using the common NGS quality control program fastqc. The second is a part of the GATK workflows, the Broad Best Practice Data Pre-processing Workflow. This workflow is intended to prepare whole-exome or whole-genome sequencing data for variant calling. The included workflow is publicly available from the Broad and Seven Bridges' public Open Workflows repository here. It has been packed into a single .cwl file for convenience. The included .yml files describe jobs that were validated through cwltool and toil.
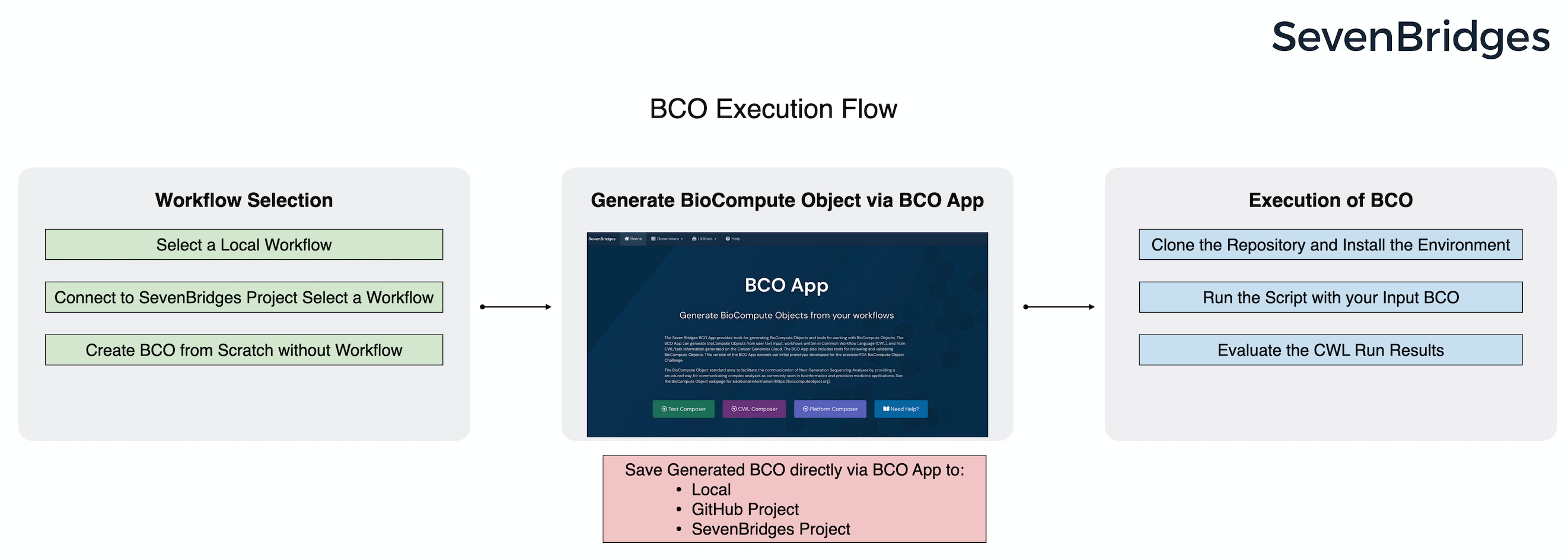
This repository include all the required apps/tools to crate BCO from scratch or CWL workflow, and provides an easy-to-use script to validate and excute given BCO with provided workflow language in its 'Execution Domain'.
Begin by cloning the repo to your local machine.
- Navigate to the BCO App Github Page: https://github.com/sbg/bco-app
- Clone the repo, or follow the directions for running through the Docker container
Using the Docker version is recommended unless doing development work on the BCO App itself.
BCO Runner Script Installation [1]
Clone the repository and install the enviroment:
git clone https://github.com/skoc/BCO-Execution.git
cd BCO-Execution/bco_runner
Create a conda environment from it as follows:
conda env create -f environment.yml
conda activate bco
python bco_runner.py --help
positional arguments:
{functions,license,validate,run_cwl}
functions list all available functions
license Prints BCO License
validate Validation options. Used to test a BCO against a JSON
schema. If no schema is supplied the ieee-2791-schema
is used as the default
run_cwl run a CWL
optional arguments:
-h, --help show this help message and exit
-v, --version show program's version number and exit
python bco_runner.py validate -b <BCO_FILE>
python bco_runner.py run_cwl -b <BCO_FILE>