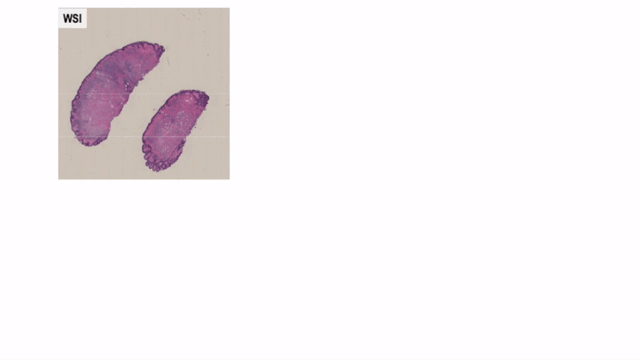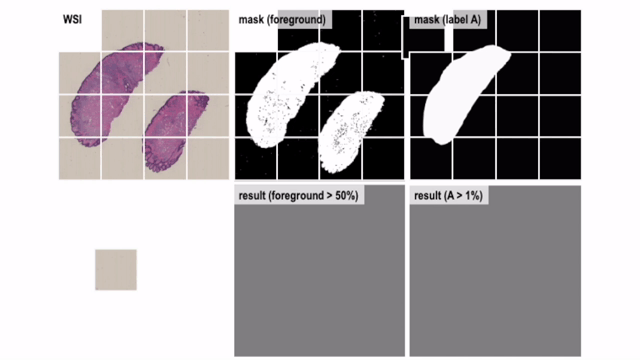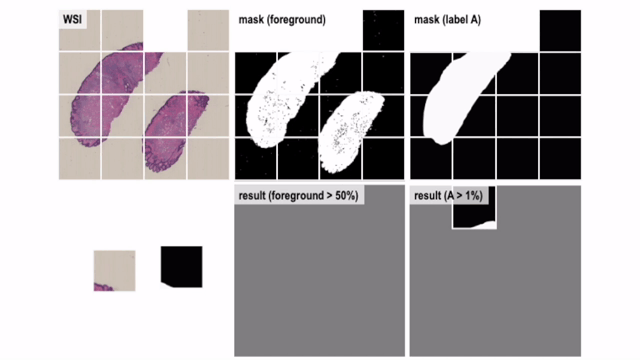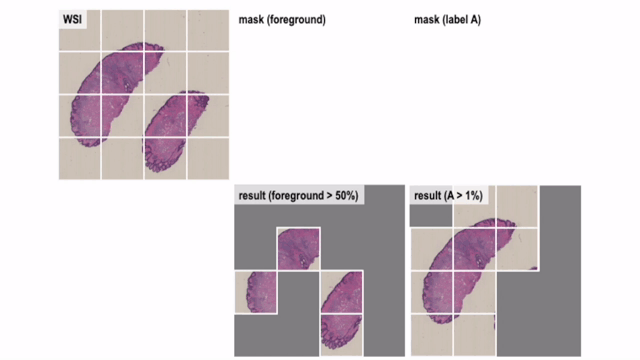
Convert Helper for Histopathological / Cytopathological Machine Learning Tasks
- Scan some WSIs.
- Make some annotations with WSI annotation tools. (ASAP and SlideRunner v.1.31.0, QuPath v0.2.3 are now available. See wiki for details.)
- Then wsiprocess helps converting WSI + Annotation data into patches and easy-to-use annotation data.
WSIPatcher will give you GUI.
See Wiki for
- available applications for annotation,
- speed comparison between patched images and loading from raw WSIs,
- how to use the other annotatiion files.
-
Install openslide or libvips. See [wiki] for installation hints.
-
Install wsiprocess
pip install wsiprocess
# Only for python 3.6 or higher
conda install -c tand826 wsiprocess
- see examples
- see wsiprocess/cli.py to check the detailed flow.
- see examples.
- ASAP
- SlideRunner version 1.31.0
- QuPath v0.2.3
- NDP.View2
- and your favorite tools with custom parser
details: wiki
Test ongoing
-
From below we tested wsi data.
- 😄 => worked well.
- ☔ => did not work well.
- otherwise => did not check
-
Aperio
- CMU-1-JP2K-33005.svs
- 😄 CMU-1-Small-Region.svs
- 😄 CMU-1.svs
- CMU-2.svs
- CMU-3.svs
- JP2K-33003-1.svs
- JP2K-33003-2.svs
-
Generic-TIFF
- ☔CMU-1.tiff
- Can not set magnification.
- ☔CMU-1.tiff
-
Hamamatsu-vms
- 😄CMU-1.zip
- CMU-2.zip
- CMU-3.zip
- Could not DOWNLOAD from http://openslide.cs.cmu.edu/download/openslide-testdata/Hamamatsu-vms/
-
Hamamatsu
- 😄CMU-1.ndpi
- CMU-2.ndpi
- CMU-3.ndpi
- OS-1.ndpi
- OS-2.ndpi
- OS-3.ndpi
-
Leica
- 😄Leica-1.scn
- Leica-2.scn
- Leica-3.scn
- Leica-Fluorescence-1.scn
-
Mirax
- CMU-1-Exported.zip
- CMU-1-Saved-1_16.zip
- CMU-1-Saved-1_2.zip
- ☔CMU-1.zip
- Can not make the foreground mask.
- CMU-2.zip
- CMU-3.zip
- Mirax2-Fluorescence-1.zip
- Mirax2-Fluorescence-2.zip
- Mirax2.2-1.zip
- Mirax2.2-2.zip
- Mirax2.2-3.zip
- Mirax2.2-4-BMP.zip
- Mirax2.2-4-PNG.zip
-
Olympus
- OS-1.zip
- OS-2.zip
- OS-3.zip
-
Trestle
- ☔CMU-1.zip
- ASAP can not show the image properly, and it's hard to annotate.
- CMU-2.zip
- CMU-3.zip
- ☔CMU-1.zip
-
Ventana
- OS-1.bif
- OS-2.bif
-
☔Zeiss : Can not load slide - ☔Zeiss-1-Merged.zvi - ☔Zeiss-1-Stacked.zvi - ☔Zeiss-2-Merged.zvi - ☔Zeiss-2-Stacked.zvi - ☔Zeiss-3-Mosaic.zvi - ☔Zeiss-4-Mosaic.zvi
curl -O -C - https://data.cytomine.coop/open/openslide/hamamatsu-ndpi/CMU-1.ndpi
- Install ASAP ( Linux / Windows ) - https://github.com/computationalpathologygroup/ASAP/releases
- Open CMU-1.ndpi and make some random annotation. - Save the annotation xml as "CMU-1.xml".
cd tests
pytest tests.py
@software{takumi_ando_2022_5938308,
author = {Takumi Ando},
title = {tand826/wsiprocess: version 0.9},
month = feb,
year = 2022,
publisher = {Zenodo},
version = {v0.9},
doi = {10.5281/zenodo.5938308},
url = {https://doi.org/10.5281/zenodo.5938308}
}