-
Notifications
You must be signed in to change notification settings - Fork 3
2.6. PCR bias correction
This tab allow you to perform PCR bias correction.
After downloading the template, fill DNA methylation values of the methylated control DNA samples into the Expectation column and upload it to shiny server.
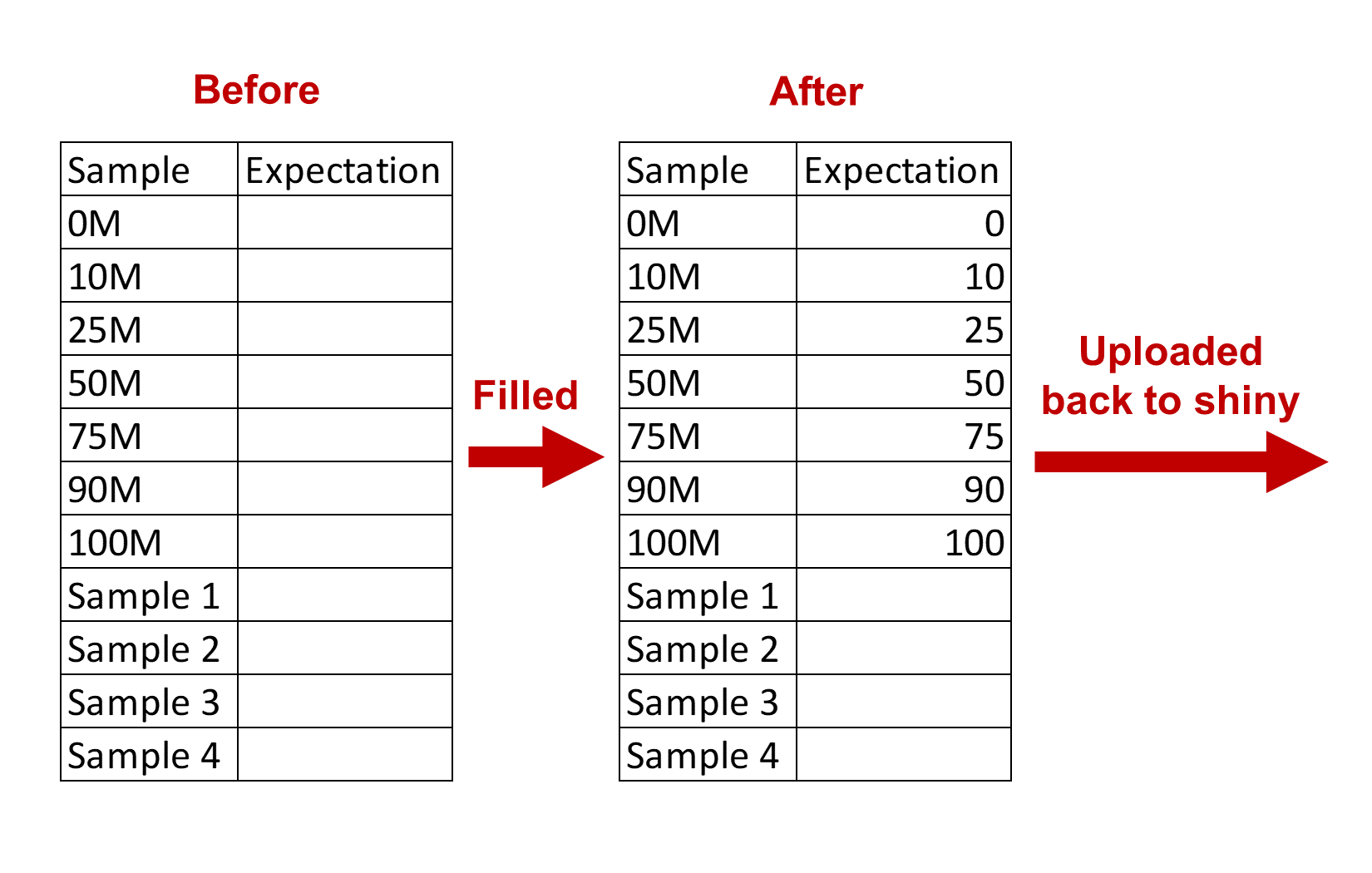
An example is available to download here.
MethPanel allows users to correct PCR bias by amplicon and CpG site. Click the download button to download a full table that contains all DNA methylation values before and after correction.
PCR-bias values are computed based on the equation:
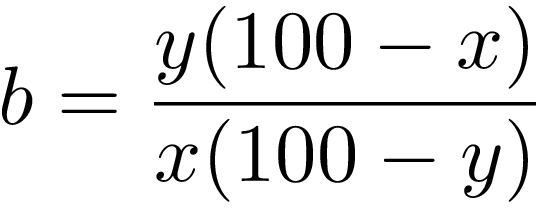 [1]
[1]
x corresponding to the expected value and y is the proportion of methylated DNA observed in methylated control DNA samples.
With the assumption the uncorrected curve is hyperbolic curve, DNA methylation is corrected as:
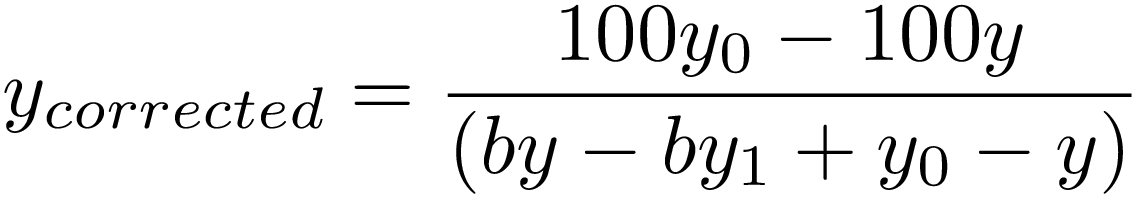 [2]
[2]
and
are the apparent methylation degree of DNA with minimum and maximum methylation, respectively.
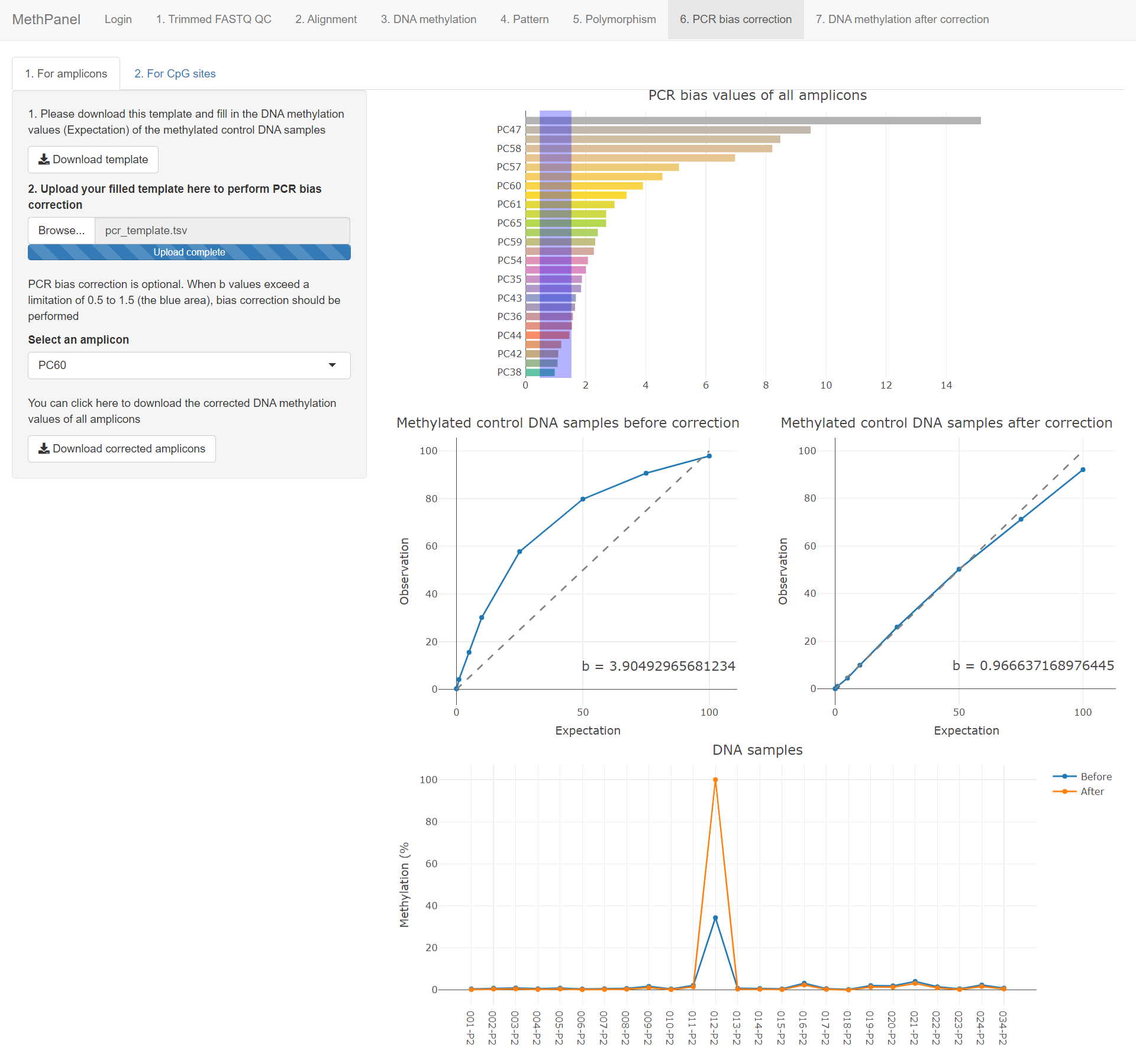
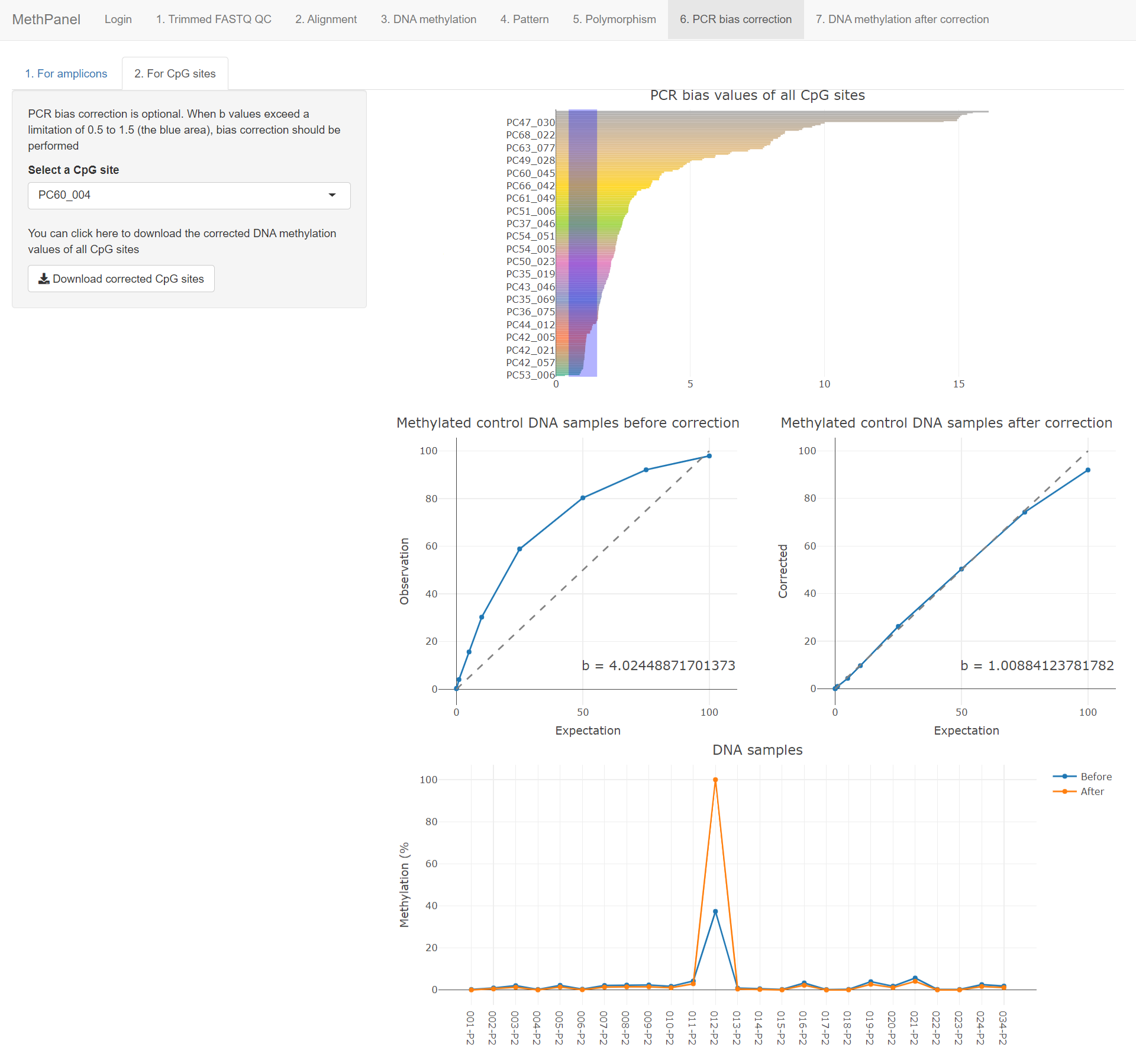
PCR bias correction is optional. The bias values (b values) are plotted in the first graph, when b values exceed a limitation of 0.5 to 1.5 (the blue area) then bias correction should be performed.
The efficiency of the correction algorithm is shown by comparing b value before and after correction of the methylated control DNA samples in the next two graphs. The last graph shows DNA methylation values of samples before and after correction.
Click the download button to download the full table that contains all DNA methylation values before and after correction.
All features are the same as in the amplicon correction, except that the bias calculation and correction was performed on individual CpG sites.
References
-
Warnecke PM, Stirzaker C, Melki JR, Millar DS, Paul CL, Clark SJ: Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic acids research 1997, 25:4422-4426.
-
Moskalev EA, Zavgorodnij MG, Majorova SP, Vorobjev IA, Jandaghi P, Bure IV, Hoheisel JD: Correction of PCR-bias in quantitative DNA methylation studies by means of cubic polynomial regression. Nucleic acids research 2011, 39:e77-e77.