-
Notifications
You must be signed in to change notification settings - Fork 44
Home
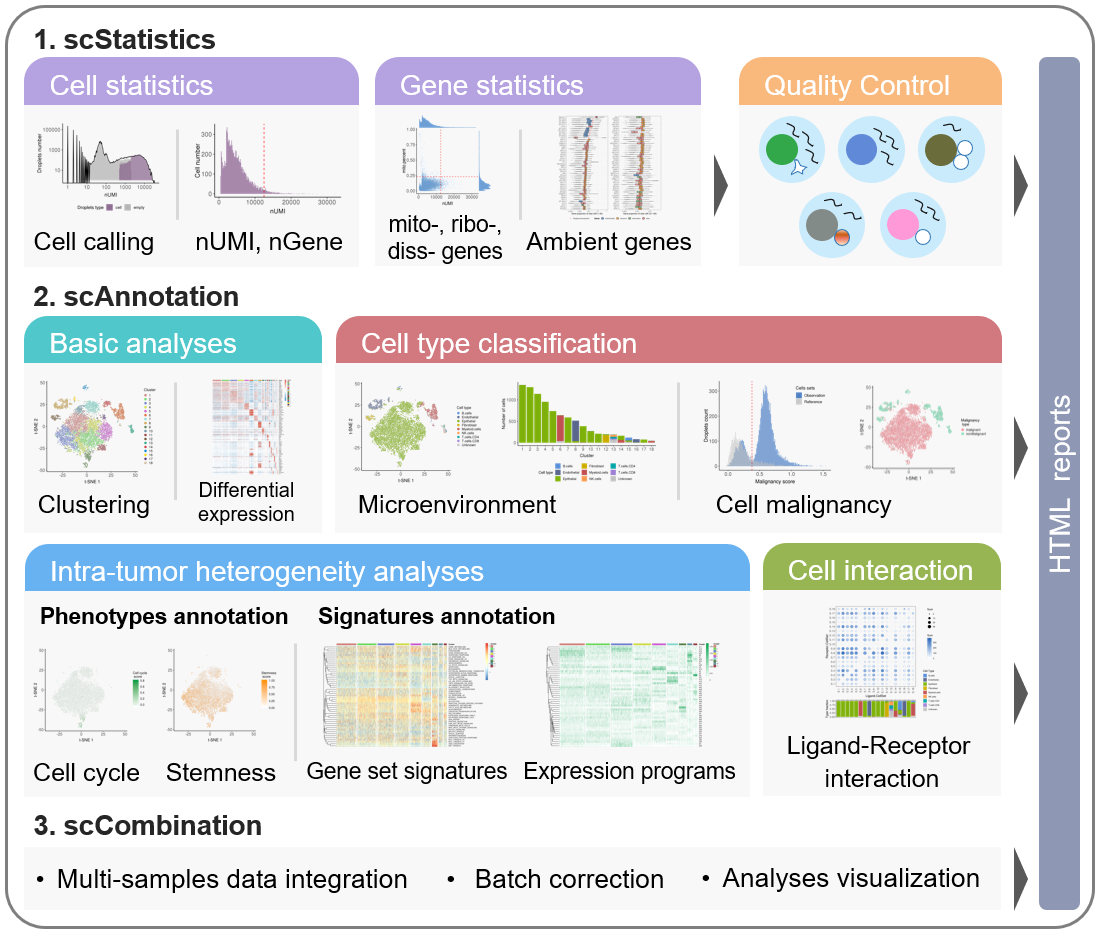
The scCancer package focuses on processing and analyzing droplet-based scRNA-seq data for cancer research. Except basic data processing steps, this package takes several special considerations for cancer-specific features.
The workflow of scCancer mainly consists of two parts: scStatistics and scAnnotation.
- The
scStatisticsperforms basic statistical analysis of raw data and quality control. - The
scAnnotationperforms functional data analyses and visualizations, such as low dimensional representation, clustering, cell type classification, malignancy estimation, cellular phenotype scoring, gene signature analysis, etc.
After these analyses, user-friendly graphic reports will be generated.
- Memery: >= 16G (for a data with ~10000 cells)
- R version: >= 3.4.0
Please install or update the package devtools by running install.packages("devtools") firstly.
Then the scCancer can be installed via
library(devtools)
devtools::install_github("wguo-research/scCancer")The vignette of scCancer can be found in the project wiki.
We also provide a kidney cancer example data, and here are the generated reports: report-scStat.html, report-scAnno.html.
Please use the following citation:
GPL-3 © G-Lab, Tsinghua University